Spindle checkpoint requires Mad1-bound and Mad1-free Mad2 - PubMed (original) (raw)
Spindle checkpoint requires Mad1-bound and Mad1-free Mad2
Eunah Chung et al. Mol Biol Cell. 2002 May.
Free PMC article
Abstract
The spindle checkpoint prevents anaphase from occurring until all chromosomes have attached properly to the mitotic spindle. The checkpoint components Mad1 and Mad2 associate with unattached kinetochores and are probably involved in triggering the checkpoint. We now demonstrate that in Xenopus egg extracts Mad1 and Mad2 form a stable complex, whereas a fraction of Mad2 molecules is not bound to Mad1. The checkpoint establishment and maintenance are lost upon titrating out free Mad2 with an excess of Mad1 or a truncated Mad1 (amino acids 326-718, Mad1C) that contains the Mad2-binding region. Mad1N (amino acids 1-445) that binds kinetochores, but not Mad2, reduces Mad1 and Mad2 at kinetochores and abolishes checkpoint maintenance. Furthermore, the association between Mad2 and Cdc20, the activator for the anaphase-promoting complex, is enhanced under checkpoint-active condition compared with that at metaphase. Immunodepletion analysis shows that the Mad1-free Mad2 protein is unable to bind Cdc20, consistent with the model that kinetochore localization of Mad2 facilitates the formation of Mad2-Cdc20 complex. This study demonstrates that the ratio between Mad1 and Mad2 is critical for maintaining a pool of Mad1-free Mad2 that is necessary for the spindle checkpoint. We propose that Mad2 may become activated and dissociated from Mad1 at kinetochores and is replenished by the pool of Mad1-free Mad2.
Figures
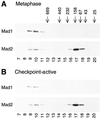
Figure 1
Gel filtration analysis of Mad1 and Mad2. Metaphase egg extract (A) or spindle checkpoint-active extract (B) was fractionated on Superose 6 column and fractions were immunoblotted for Mad1 (top) or Mad2 (bottom). The fraction number is indicated on the bottom. The fractionation of size standards is indicated on top.
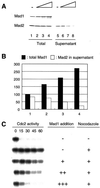
Figure 2
Excess Mad1 titrates out Mad1-free Mad2 and abolishes spindle checkpoint. (A) Immunoblot analysis of Mad1 and Mad2 in extracts containing increasing amount of Mad1 translation. The samples were immunoprecipitated with anti-Mad1 antibodies. Total proteins before immunoprecipitation (lanes 1–4) or supernatants left after immunoprecipitation (lanes 5–8) were analyzed by immunoblot with anti-Mad1 or anti-Mad2 antibodies as indicated. The level of Mad2 left in the supernatant decreases with increasing amount of Mad1 added. Mock translation was added, so that all samples contained the same amount of translation reaction. A graph of the result is shown in B. The relative level of total Mad1 (lanes 1–4 in A) and Mad2 in the supernatant (lanes 5–8 in A) is shown. The amount of Mad1 and Mad2 in lane 1 of A is designated as 100. (C) Autoradiograms of histone H1 kinase assay. The same samples shown in lanes 1–4 of A were incubated with sperm nuclei and nocodazole for 30 min. Calcium chloride was then added to trigger mitotic exit. Samples were taken every 15 min for histone H1 kinase assay as indicated on top. In the presence of excess Mad1, the extract failed to activate the checkpoint.
Figure 3
Addition of Mad2 reverses the effect of excess Mad1 and restores the checkpoint. Extracts were incubated with nuclei and nocodazole in the presence of Mad1 translation alone, Mad2 translation alone, or both. Samples were processed for immunoblot analysis (A) and for H1 kinase measurement (B) as described in Figure 2. Restoration of the checkpoint by the addition of Mad2 indicates that the ratio between Mad1 and Mad2 levels is critical.
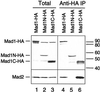
Figure 4
Immunoblot analysis of truncated Mad1 proteins and their interaction with Mad2. Extracts containing translation of HA-tagged full-length Mad1 (lanes 1 and 4), amino acids 1–445 (Mad1N, lanes 2 and 5), or amino acids 326–718 (Mad1C, lanes 3 and 6) were immunoprecipitated with anti-HA antibodies. Total proteins in the extracts (lanes 1–3) and the proteins associated with the immunoprecipitates (lanes 4–6) were analyzed by immunoblot with anti-Mad1 (top) or anti-Mad2 (bottom) antibodies. Both full-length Mad1 and Mad1C coimmunoprecipitate with Mad2. No self-interaction of Mad1 is detected. The migration of various HA-tagged Mad1 proteins is indicated on the left, and molecular size standards are indicated on the right.
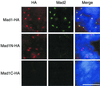
Figure 5
Kinetochore binding domain resides within amino acids 1–445 of Mad1. Mitotic extracts were immunodepleted with anti-Mad1 antibodies to remove endogenous Mad1. The extracts were supplemented with HA-tagged full-length Mad1, Mad1N, or Mad1C that were translated in Mad1-depleted extracts. After incubating the extracts with sperm nuclei and nocodazole, chromosomes were isolated and stained with mouse anti-HA and rabbit anti-Mad2 antibodies. Fluorescein-conjugated anti-rabbit and Texas Red-conjugated anti-mouse IgG antibodies were used as secondary antibodies. The DNA was stained with the DNA-binding dye Hoechst 33258. The merges of all three fluorochromes are also shown (Merge). All pictures of the same fluorochrome were processed in the same way. Both full-length Mad1 and Mad1N associate with kinetochores. Similar results were obtained in the absence of Mad2 (our unpublished data). Bar, 10 μm.
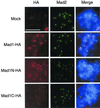
Figure 6
Mad1N is recruited onto kinetochores that have bound with endogenous Mad1 and Mad2. Sperm nuclei and nocodazole were incubated in mitotic extract for 30 min followed by the addition of mock, full-length Mad1, Mad1N, or Mad1C translated in extract depleted for both Mad1 and Mad2. Samples were incubated for another 30 min and then processed for immunofluorescence staining as described for Figure 5.
Figure 7
Mad2 levels in the chromosomal fractions are reduced by additional Mad1N, Mad1C, and Mad1. (A) Coomassie staining of proteins associated with chromosomes that were isolated from extracts treated with (lane 2) or without (lane 1) nocodazole. The migration of molecular weight standards is indicated on the left. The predominant proteins are labeled on the right. (B) Immunoblot of total proteins or chromosomal fractions with anti-Mad1 or anti-Mad2 antibodies. As described for Figure 6, mitotic chromosomes were assembled in extracts with (lanes 2–5 and 7–10) or without (lanes 1 and 6) nocodazole, followed by the addition of mock, Mad1N, Mad1C, or Mad1 translation as indicated on top. Chromosomes were isolated and analyzed by immunoblot for Mad1 (top) or Mad2 (bottom). The addition of Mad1N reduces chromosomal Mad2 to 43% of that on mock-challenged chromosomes. Those challenged with Mad1C or Mad1 contained ∼70% of Mad2.
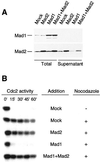
Figure 8
Mad1N and Mad1C abolish checkpoint establishment and maintenance. (A) Autoradiograms of histone H1 kinase assay. For checkpoint establishment samples, the translation of mock, Mad1N, or Mad1C was added to CSF-arrested extract, followed by incubation with sperm nuclei and nocodazole for 20 min to activate the spindle checkpoint. For checkpoint maintenance, the checkpoint was first activated in CSF-arrested extract by incubation with nuclei and nocodazole for 20 min, followed by the addition of translation and another incubation for 30 min. Calcium chloride was then added to trigger mitotic exit. Samples were taken immediately before the addition of calcium chloride (time 0) and every 15 min thereafter for histone H1 kinase assay as indicated on top. (B) Immunoblot analysis of Mad1, Mad1N, Mad1C, and Mad2 in samples used in A.
Figure 9
Unattached chromosomes enhance Mad2–Cdc20 interaction. (A) Spindle checkpoint enhances Mad2–Cdc20 interaction. Interphase (lanes 1 and 4), CSF-arrested (metaphase, lanes 2 and 5), or spindle checkpoint-active (lanes 3, 6, and 7) extracts containing the same number of nuclei were subjected to immunoprecipitation with anti-Cdc20 antibody (lanes 4–6) or with a control antibody (lane 7). Total protein in the extract (lanes 1–3) or the immunoprecipitates (lanes 4–7) was then immunoblotted for Cdc20 (top) or Mad2 (bottom). The band below Cdc20 in lanes 4–7 is the IgG heavy chain. (B) Cdc20 is the limiting factor in the complex of Mad2–Cdc20. Extracts containing increasing amounts of Cdc20 translation as indicated on top were incubated with sperm nuclei and nocodazole to activate the spindle checkpoint, followed by immunoprecipitation with anti-Cdc20 antibody. Total protein in the extract (lanes 1–3) or the immunoprecipitates (lanes 4–6) was then immunoblotted for Cdc20 (top) or Mad2 (bottom). Cdc20 in lane 1 is at the endogenous level. (C) Preventing kinetochore binding of Mad2 abolishes Mad2–Cdc20 interaction. CSF-arrested extract was immunodepleted with control antibody (lane 1), with anti-Mad2 antibody (lane 3), or with anti-Mad1 antibody (lane 4). Mad2 depletion removed both Mad2 and Mad1, whereas Mad1 depletion left behind >50% of Mad2. Lane 2 is a mix of mock- and Mad2-depleted extract (1:1), which contained 50% of Mad1 and Mad2. After incubation with sperm nuclei and nocodazole, the extracts were subjected to anti-Cdc20 immunoprecipitation, followed by immunoblot for Cdc20 or Mad2 as indicated (bottom two panels). Proteins in the extracts were also immunoblotted for Cdc20, Mad2, or Mad1 as indicated on the left (top three panels). (D) Level of Mad2–Cdc20 interaction is proportional to the number of unattached chromosomes. Extracts were incubated with nocodazole and sperm nuclei at density indicated on top (per microliter of extract), followed by Cdc20 immunoprecipitation and immunoblot analysis. The amount of Mad2 associated with Cdc20 levels off at the nuclear density of 8,000–16,000.
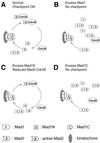
Figure 10
Models for how Mad1N and Mad1C may act as dominant negatives for the spindle checkpoint through different mechanisms. (A) Under normal condition, Mad1 targets its associated Mad2 to kinetochores. The pool of Mad1-free Mad2 protein ensures that Mad1 quickly binds new Mad2 when active Mad2 or Mad2–Cdc20 complex has dissociated, thus maintaining the checkpoint signal. (B and D) Excess Mad1 or Mad1C reduces the pool of Mad1-free Mad2 molecules, so that the kinetochores are not replenished with Mad2 once the previous Mad2 has left. (C) Mad1N replaces the full-length Mad1 on kinetochores. It fails to recruit Mad2, thus competing with wild-type Mad1 and Mad2 in binding kinetochores.
Similar articles
- BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1.
Chen RH. Chen RH. J Cell Biol. 2002 Aug 5;158(3):487-96. doi: 10.1083/jcb.200204048. Epub 2002 Aug 5. J Cell Biol. 2002. PMID: 12163471 Free PMC article. - Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity.
Sharp-Baker H, Chen RH. Sharp-Baker H, et al. J Cell Biol. 2001 Jun 11;153(6):1239-50. doi: 10.1083/jcb.153.6.1239. J Cell Biol. 2001. PMID: 11402067 Free PMC article. - Spindle checkpoint protein dynamics at kinetochores in living cells.
Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Howell BJ, et al. Curr Biol. 2004 Jun 8;14(11):953-64. doi: 10.1016/j.cub.2004.05.053. Curr Biol. 2004. PMID: 15182668 - Checkpoint signalling: Mad2 conformers and signal propagation.
Hardwick KG. Hardwick KG. Curr Biol. 2005 Feb 22;15(4):R122-4. doi: 10.1016/j.cub.2005.02.008. Curr Biol. 2005. PMID: 15723780 Review. - Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model.
Yu H. Yu H. J Cell Biol. 2006 Apr 24;173(2):153-7. doi: 10.1083/jcb.200601172. J Cell Biol. 2006. PMID: 16636141 Free PMC article. Review.
Cited by
- Septin 9 controls CCNB1 stabilization via APC/CCDC20 during meiotic metaphase I/anaphase I transition in mouse oocytes.
Chen L, Ouyang YC, Gu LJ, Guo JN, Han ZM, Wang ZB, Hou Y, Schatten H, Sun QY. Chen L, et al. Cell Prolif. 2023 Feb;56(2):e13359. doi: 10.1111/cpr.13359. Epub 2022 Nov 10. Cell Prolif. 2023. PMID: 36354207 Free PMC article. - Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis.
Bentley AM, Normand G, Hoyt J, King RW. Bentley AM, et al. Mol Biol Cell. 2007 Dec;18(12):4847-58. doi: 10.1091/mbc.e06-06-0539. Epub 2007 Sep 19. Mol Biol Cell. 2007. PMID: 17881737 Free PMC article. - Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension.
Shannon KB, Canman JC, Salmon ED. Shannon KB, et al. Mol Biol Cell. 2002 Oct;13(10):3706-19. doi: 10.1091/mbc.e02-03-0137. Mol Biol Cell. 2002. PMID: 12388768 Free PMC article. - Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator.
Lince-Faria M, Maffini S, Orr B, Ding Y, Cláudia Florindo, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Lince-Faria M, et al. J Cell Biol. 2009 Mar 9;184(5):647-57. doi: 10.1083/jcb.200811012. J Cell Biol. 2009. PMID: 19273613 Free PMC article. - Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting.
Kim S, Sun H, Tomchick DR, Yu H, Luo X. Kim S, et al. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6549-54. doi: 10.1073/pnas.1118210109. Epub 2012 Apr 9. Proc Natl Acad Sci U S A. 2012. PMID: 22493223 Free PMC article.
References
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe J. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources