The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase - PubMed (original) (raw)
The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase
Thomas B Broudy et al. Infect Immun. 2002 Jun.
Abstract
The role lysogenic bacteriophage play in the pathogenesis of the host bacterium is poorly understood. In a previous study, we found that streptococcal coculture with human pharyngeal cells resulted in the induction of lysogenic bacteriophage as well as the phage-associated streptococcal pyrogenic exotoxin C (SpeC). In this study, we have determined that in addition to SpeC induction, a number of other streptococcal proteins are also released by the bacteria during coculture with pharyngeal cells. Among these, we identified and characterized a novel 27-kDa secreted protein. Sequence analysis of this novel protein demonstrated it to be encoded by the same lysogenic bacteriophage which harbors speC. Protein sequence analysis revealed varied homologies with several streptococcal DNases. Further biochemical characterization of the recombinantly expressed protein verified it to be a divalent cation-dependent streptococcal phage-encoded DNase (Spd1). Although functionally distinct, SpeC and Spd1 are associated by a number of parameters, including genetic proximity and transcriptional regulation. Finally, we speculate on the induction of phage-encoded DNase (Spd1) enhancing the fitness of both bacteria and phage.
Figures
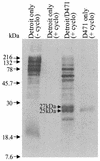
FIG. 1.
[35S]Met-labeled proteins in the culture media of the non-cycloheximide-treated pharyngeal-cell control [Detroit only (− cyclo)] and the cycloheximide-treated samples, including the pharyngeal-cell control [Detroit only (+ cyclo)], pharyngeal-cell-S. pyogenes coculture (Detroit/D471), and S. pyogenes control (D471 only).
FIG. 2.
Schematic alignment of spd1 and speC sequences from the integrated prophage in the M1 genome and the region of the M6 strain D471 which was sequenced. attL and attR, left and right attachment sites, respectively, of the integrated prophage.
FIG. 3.
Southern blot of _Hin_dIII-digested genomic DNAs from 19 strains of S. pyogenes probed with either spd1 or speC. The M serotype is designated in parentheses next to the strain number, and the strains are grouped by their surface-exposed antigenic properties as either class I or class II. The DNA ladder is shown to the left, with arrows indicating 1-kb increments.
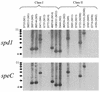
FIG. 4.
RNAs extracted from Detroit 562 cells, D471 cocultured with Detroit 562 (Cocultured D471), and D471 alone were blotted and probed for either spd1 or speC.
FIG. 5.
Culture supernatants from S. gordonii (GP251) parent strain and supernatants from recombinant S. gordonii (secreting either NucA or Spd1) were collected. Each supernatant was incubated with pBlueScript DNA in Tris-Ca2+-Mg2+ buffer, either with (+) or without (−) EDTA present. The completed reaction was analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining.
FIG. 6.
(A) Amino acid sequence alignment of the Spd1 nuclease with the known nuclease MF/DNase B and three other putative DNases, including two from S. pyogenes (MF2 and MF3) and one from L. lactis (YbfB). The conserved histidine is indicated with an arrowhead. The dots represent spaces in the sequence alignment. Solid boxes represent amino acid identity, and shaded boxes represent amino acid similarity based on Clustal W sequence analysis. (B) Amino acid sequence identities between MF/DNase B or Spd1 and the Spd1-related putative DNases.
FIG. 6.
(A) Amino acid sequence alignment of the Spd1 nuclease with the known nuclease MF/DNase B and three other putative DNases, including two from S. pyogenes (MF2 and MF3) and one from L. lactis (YbfB). The conserved histidine is indicated with an arrowhead. The dots represent spaces in the sequence alignment. Solid boxes represent amino acid identity, and shaded boxes represent amino acid similarity based on Clustal W sequence analysis. (B) Amino acid sequence identities between MF/DNase B or Spd1 and the Spd1-related putative DNases.
Similar articles
- Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells.
Broudy TB, Pancholi V, Fischetti VA. Broudy TB, et al. Infect Immun. 2001 Mar;69(3):1440-3. doi: 10.1128/IAI.69.3.1440-1443.2001. Infect Immun. 2001. PMID: 11179310 Free PMC article. - Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes.
Aziz RK, Ismail SA, Park HW, Kotb M. Aziz RK, et al. Mol Microbiol. 2004 Oct;54(1):184-97. doi: 10.1111/j.1365-2958.2004.04255.x. Mol Microbiol. 2004. PMID: 15458415 - The structural characterization of a prophage-encoded extracellular DNase from Streptococcus pyogenes.
Korczynska JE, Turkenburg JP, Taylor EJ. Korczynska JE, et al. Nucleic Acids Res. 2012 Jan;40(2):928-38. doi: 10.1093/nar/gkr789. Epub 2011 Sep 24. Nucleic Acids Res. 2012. PMID: 21948797 Free PMC article. - The DNases of pathogenic Lancefield streptococci.
Remmington A, Turner CE. Remmington A, et al. Microbiology (Reading). 2018 Mar;164(3):242-250. doi: 10.1099/mic.0.000612. Epub 2018 Jan 25. Microbiology (Reading). 2018. PMID: 29458565 Review. - The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence.
Banks DJ, Beres SB, Musser JM. Banks DJ, et al. Trends Microbiol. 2002 Nov;10(11):515-21. doi: 10.1016/s0966-842x(02)02461-7. Trends Microbiol. 2002. PMID: 12419616 Review.
Cited by
- In vivo lysogenic conversion of Tox(-) Streptococcus pyogenes to Tox(+) with Lysogenic Streptococci or free phage.
Broudy TB, Fischetti VA. Broudy TB, et al. Infect Immun. 2003 Jul;71(7):3782-6. doi: 10.1128/IAI.71.7.3782-3786.2003. Infect Immun. 2003. PMID: 12819060 Free PMC article. - The Bacteriophages of Streptococcus pyogenes.
McShan WM, McCullor KA, Nguyen SV. McShan WM, et al. Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0059-2018. doi: 10.1128/microbiolspec.GPP3-0059-2018. Microbiol Spectr. 2019. PMID: 31111820 Free PMC article. Review. - Novel algorithms reveal streptococcal transcriptomes and clues about undefined genes.
Ryan PA, Kirk BW, Euler CW, Schuch R, Fischetti VA. Ryan PA, et al. PLoS Comput Biol. 2007 Jul;3(7):e132. doi: 10.1371/journal.pcbi.0030132. PLoS Comput Biol. 2007. PMID: 17616984 Free PMC article. - Lysogenic transfer of group A Streptococcus superantigen gene among Streptococci.
Vojtek I, Pirzada ZA, Henriques-Normark B, Mastny M, Janapatla RP, Charpentier E. Vojtek I, et al. J Infect Dis. 2008 Jan 15;197(2):225-34. doi: 10.1086/524687. J Infect Dis. 2008. PMID: 18179387 Free PMC article. - The prophage-encoded hyaluronate lyase has broad substrate specificity and is regulated by the N-terminal domain.
Singh SK, Bharati AP, Singh N, Pandey P, Joshi P, Singh K, Mitra K, Gayen JR, Sarkar J, Akhtar MS. Singh SK, et al. J Biol Chem. 2014 Dec 19;289(51):35225-36. doi: 10.1074/jbc.M113.507673. Epub 2014 Nov 6. J Biol Chem. 2014. PMID: 25378402 Free PMC article.
References
- Alouf, J. E. 1986. Streptococcal toxins (streptolysin O, streptolysin S, erythrogenic toxin), p. 635-691. In F. Dorner and J. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, New York, N.Y. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources