The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling - PubMed (original) (raw)
The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling
Roberta Roncarati et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The beta-amyloid precursor protein (APP) and the Notch receptor undergo intramembranous proteolysis by the Presenilin-dependent gamma-secretase. The cleavage of APP by gamma-secretase releases amyloid-beta peptides, which have been implicated in the pathogenesis of Alzheimer's disease, and the APP intracellular domain (AID), for which the function is not yet well understood. A similar gamma-secretase-mediated cleavage of the Notch receptor liberates the Notch intracellular domain (NICD). NICD translocates to the nucleus and activates the transcription of genes that regulate the generation, differentiation, and survival of neuronal cells. Hence, some of the effects of APP signaling and Alzheimer's disease pathology may be mediated by the interaction of APP and Notch. Here, we show that membrane-tethered APP binds to the cytosolic Notch inhibitors Numb and Numb-like in mouse brain lysates. AID also binds Numb and Numb-like, and represses Notch activity when released by APP. Thus, gamma-secretase may have opposing effects on Notch signaling; positive by cleaving Notch and generating NICD, and negative by processing APP and generating AID, which inhibits the function of NICD.
Figures
Figure 1
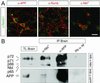
APP and Numbs/Numb-like are coexpressed with Notch1 in cortical neurons and are coimmunoprecipitated from adult mouse brain. (A) Representative confocal images (1 μm thick) of cultured cortical neurons immunostained for APP, Numb, Numb-like (all red), and Notch1 (green). (Bar = 15 μm.) (B) Coimmunoprecipitation (IP) of APP and Nbl from total lysates (TL) of adult mouse brain was done by using the α-APP (no. 1736) and α-Nbl antibodies. *, α-Nbl polyclonal antibody crossreacts with all four Numb isoforms. Western blotting (WB) was performed with either the α-Nbl antiserum or the α-APP monoclonal antibody 22C11. The rabbit α-mouse IgG (Rb α-M) antibody was used as a negative control.
Figure 2
AID interacts with Nbl and Numbs. (A) HEK293 cells were transfected with either AID-ECFP or ECFP along with Flag-Numb p72 (p72, Upper), Flag-Numb p71 (p71, Upper middle), Flag-Numb p66 (p66, Lower middle), or Flag-Numb p65 (p65, Lower). Cell lysates were coimmunoprecipitated (IP) with either α-Flag or α-Living Colors (α-LC), which recognizes ECFP- and EYFP-tagged proteins. Expression of transfected constructs in total lysates (TL) and immunoprecipitates (IP) also were analyzed by Western blot (WB) with the α-Nbl or α-LC antibodies. p72ΔN, p71ΔN, p66ΔN, and p65ΔN are recognized by the α-Nbl antisera, which is specific for the COOH-terminal region of Numb/Nbl, but are not immunoprecipitated by the α-Flag antibody, which is specific for the Flag epitope fused at the NH2 terminus of these proteins. Therefore, they represent degradation products lacking the NH2-terminal portion and part of the PTB domain, which do not interact with AID-ECFP. (B) HEK293 cells were transfected with AID-ECFP or ECFP along with either Flag-Nbl (lanes 1–6) or the unrelated protein Flag-AIP (lanes 7–12). Lysates were immunoprecipitated with either the α-Flag or α-LC antibodies as indicated. Lysates and immunoprecipitates were analyzed by WB with either the α-Flag, α-LC, or α-Nbl antibodies as indicated. NblΔN is a degradation product of Nbl lacking the NH2-terminal PTB domain and does not interact with AID-ECFP (see above for explanation). * indicates degradation products of AID fusion proteins containing ECFP alone, which does not bind Nbl. (C) HEK293 cells were transfected with Flag-Nbl along with EYFP-tagged deletion construction of AID as indicated. Lysates were immunoprecipitated with α-Flag and analyzed by WB with either α-Nbl or α-LC as indicated. AID mutant numbering is based on the AID59 peptide sequence. (D) HEK 293 cells were transfected with either AID-EYFP or 12–28AID-EYFP along with Flag tagged deletion constructs of Nbl as indicated. Lysates were immunoprecipitated with α-Flag and analyzed by WB with either α-Flag or α-LC as indicated. ** indicates the signal from the α-Flag heavy chain. The difference in molecular mass between AID-EYFP and mutant forms of AID-EYFP, which can be appreciated in (C), is not apparent in (D), because the proteins were separated less extensively by SDS/PAGE.
Figure 3
Nbl interacts with AID and membrane-tethered APP in vivo. (A) Representative plot of ECFP vs. EYFP. The R16 gate, containing cells with nearly equal ECFP and EYFP intensity, was used to calculate the percentage of cotransfected cells exhibiting FRET. (B) HEK293 cells were transfected with the indicated EYFP-tagged APP/AID constructs (Left) along with ECFP-Nbl constructs (Right). Samples are plotted based on the intensity of the FRET signal (y axis) versus its ECFP or EYFP emission (x axis, left and right column, respectively). Cells in the R11 (ECFP column) or R3 (EYFP column) gates were scored as positive. Percentages are calculated as the number of positive cells divided by the total number of cells, or, in parentheses, the number of cotransfected cells (gate R16). Although both APP and AID interact with Nbl (as indicated by positive FRET), the AID mutants 12–28AID-EYFP, 39–55AID-EYFP, and 44–59AID-EYFP do not exhibit FRET when coexpressed with ECFP-Nbl. In addition, the Nbl deletion mutant ECFP-Nbl212–603, which lacks the PTB domain, does not interact with AID-EYFP to detectable levels. (C) Effect of the γ-secretase inhibitor DAPT (100 nM) on FRET between Nbl and AID or APP. HEK293 cells were transfected with ECFP-Nbl along with either EYFP-AID or EYFP-APP. Inhibition of γ-secretase activity by DAPT doubles the number of ECFP-Nbl/EYFP-APP transfected cells exhibiting FRET. As expected, DAPT does not affect FRET in ECFP-Nbl/EYFP-AID transfected cells.
Figure 4
Processing of APP and release of AID inhibits Notch signaling. (A) HeLa cells were transfected with 4xCBF1-luciferase (CBF-luc) along with the indicated Notch1 and Nbl constructs. Nbl inhibits the transactivation of CBF-luc by both NICD and NΔE. (B) Western blotting (WB) with the α-Nbl antiserum shows expression of Nbl and/or Numb proteins in HeLa cells (Top). WB with α-APP (22C11) shows induction of APP by Dox in APP-HeLa Tet-on cells (Middle). An α-PARP antibody (2C10) was used to normalize for protein loading (Bottom). (C and D) APP-HeLa Tet-on cells were transfected with CBF-luc along with (C) NICD or (D) NΔE. Some samples were treated with Dox to induce APP expression and/or with the γ-secretase inhibitor DAPT (100 nM). The decrease in NICD activity by APP induction is inhibited by DAPT (C). DAPT treatment significantly reduces NΔE activity in HeLa Tet-on cells (D). (E_–_H) HeLa cells were transfected with CBF-luc along NICD, NΔE, AID, or mutant forms of AID as indicated. AID inhibits the activation of CBF-luc by both NICD and NΔE (E). The inhibition is dose-dependent (F) and correlates with the ability of AID to interact with Nbl (G). (H) HeLa cells were transfected with either CBF-luc or GAL4-luciferase (GAL4-luc) reporter genes along with APPCT-Gal4 in the presence or absence of cotransfected Fe65. APPCT-Gal4 reduces NICD activity but enhances Fe65 transactivation. Significance was determined by using a two-tailed Student's t test (*, P < 0.05; **, P ≤ 0 01; ***, P ≤ 0.001).
Figure 5
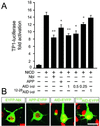
AID inhibits NICD signaling in primary cortical neurons. (A) Cultured cortical neurons (5 to 6 days old) were transfected with TP1-luciferase along with NICD, Nbl, APP, AID, or AID12–28 as indicated. The activity of NICD was inhibited by Nbl, APP, and AID in a dose-dependent manner (0.25, 0.5, and 1 μg/well). No significant change in NICD-dependent transactivation of TP1-luciferase was observed when NICD was cotransfected with AID12–28. Significance was determined by using a two-tailed Student's t test (*, P < 0.05; **, P ≤ 0.01). (B) Representative confocal images (1 μm thick) of cultured cortical neurons transiently transfected with AID-EYFP, AID12–28-EYFP, APP-EYFP, and EYFP-Nbl fusion constructs (shown in green). Neuronal DNA was stained with propidium iodide (shown in red). (Bar = 20 μm.)
Similar articles
- The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner.
Kimberly WT, Zheng JB, Guénette SY, Selkoe DJ. Kimberly WT, et al. J Biol Chem. 2001 Oct 26;276(43):40288-92. doi: 10.1074/jbc.C100447200. Epub 2001 Sep 5. J Biol Chem. 2001. PMID: 11544248 - A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain.
De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. De Strooper B, et al. Nature. 1999 Apr 8;398(6727):518-22. doi: 10.1038/19083. Nature. 1999. PMID: 10206645 - A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein.
Hass MR, Yankner BA. Hass MR, et al. J Biol Chem. 2005 Nov 4;280(44):36895-904. doi: 10.1074/jbc.M502861200. Epub 2005 Aug 15. J Biol Chem. 2005. PMID: 16103124 Free PMC article. - Implication of APP secretases in notch signaling.
Hartmann D, Tournoy J, Saftig P, Annaert W, De Strooper B. Hartmann D, et al. J Mol Neurosci. 2001 Oct;17(2):171-81. doi: 10.1385/JMN:17:2:171. J Mol Neurosci. 2001. PMID: 11816790 Review. - Role of presenilin in gamma-secretase cleavage of amyloid precursor protein.
Xia W. Xia W. Exp Gerontol. 2000 Jul;35(4):453-60. doi: 10.1016/s0531-5565(00)00111-x. Exp Gerontol. 2000. PMID: 10959033 Review.
Cited by
- Interference of human and Drosophila APP and APP-like proteins with PNS development in Drosophila.
Merdes G, Soba P, Loewer A, Bilic MV, Beyreuther K, Paro R. Merdes G, et al. EMBO J. 2004 Oct 13;23(20):4082-95. doi: 10.1038/sj.emboj.7600413. Epub 2004 Sep 23. EMBO J. 2004. PMID: 15385958 Free PMC article. - The senescence hypothesis of disease progression in Alzheimer disease: an integrated matrix of disease pathways for FAD and SAD.
Hunter S, Arendt T, Brayne C. Hunter S, et al. Mol Neurobiol. 2013 Dec;48(3):556-70. doi: 10.1007/s12035-013-8445-3. Epub 2013 Apr 3. Mol Neurobiol. 2013. PMID: 23546742 Review. - Downregulation of Hes1 expression in experimental biliary atresia and its effects on bile duct structure.
Zhang RZ, Zeng XH, Lin ZF, Ming-Fu, Tong YL, Lui VC, Tam PK, Lamb JR, Xia HM, Chen Y. Zhang RZ, et al. World J Gastroenterol. 2018 Aug 7;24(29):3260-3272. doi: 10.3748/wjg.v24.i29.3260. World J Gastroenterol. 2018. PMID: 30090006 Free PMC article. - The amyloid-beta precursor protein: integrating structure with biological function.
Reinhard C, Hébert SS, De Strooper B. Reinhard C, et al. EMBO J. 2005 Dec 7;24(23):3996-4006. doi: 10.1038/sj.emboj.7600860. Epub 2005 Oct 27. EMBO J. 2005. PMID: 16252002 Free PMC article. Review. - Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex.
Chen JG, Rasin MR, Kwan KY, Sestan N. Chen JG, et al. Proc Natl Acad Sci U S A. 2005 Dec 6;102(49):17792-7. doi: 10.1073/pnas.0509032102. Epub 2005 Nov 28. Proc Natl Acad Sci U S A. 2005. PMID: 16314561 Free PMC article.
References
- Price D L, Tanzi R E, Borchelt D R, Sisodia S S. Annu Rev Genet. 1998;32:461–493. - PubMed
- Selkoe D J. Physiol Rev. 2001;81:741–766. - PubMed
- Esler W P, Wolfe M S. Science. 2001;293:1449–1454. - PubMed
- Haass C, De Strooper B. Science. 2001;286:916–919. - PubMed
- Passer B, Pellegrini L, Russo C, Siegel R M, Lenardo M J, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimer Dis. 2000;2:289–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 DA015014/DA/NIDA NIH HHS/United States
- R01 DA015014/DA/NIDA NIH HHS/United States
- R01 AG019394/AG/NIA NIH HHS/United States
- DA15014-01/DA/NIDA NIH HHS/United States
- NS14841/NS/NINDS NIH HHS/United States
- AG19394/AG/NIA NIH HHS/United States
- R01 NS014841/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases