Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands - PubMed (original) (raw)
Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands
Karen Honey et al. J Exp Med. 2002.
Abstract
CD4+ T cells are positively selected in the thymus on peptides presented in the context of major histocompatibility complex class II molecules expressed on cortical thymic epithelial cells. Molecules regulating this peptide presentation play a role in determining the outcome of positive selection. Cathepsin L mediates invariant chain processing in cortical thymic epithelial cells, and animals of the I-A(b) haplotype deficient in this enzyme exhibit impaired CD4+ T cell selection. To determine whether the selection defect is due solely to the block in invariant chain cleavage we analyzed cathepsin L-deficient mice expressing the I-A(q) haplotype which has little dependence upon invariant chain processing for peptide presentation. Our data indicate the cathepsin L defect in CD4+ T cell selection is haplotype independent, and thus imply it is independent of invariant chain degradation. This was confirmed by analysis of I-A(b) mice deficient in both cathepsin L and invariant chain. We show that the defect in positive selection in the cathepsin L-/- thymus is specific for CD4+ T cells that can be selected in a wild-type and provide evidence that the repertoire of T cells selected differs from that in wild-type mice, suggesting cortical thymic epithelial cells in cathepsin L knockout mice express an altered peptide repertoire. Thus, we propose a novel role for cathepsin L in regulating positive selection by generating the major histocompatibility complex class II bound peptide ligands presented by cortical thymic epithelial cells.
Figures
Figure 1.
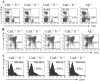
Impaired selection of CD4+ but not CD8+ T cell receptor transgenic T cells in cathepsin L–deficient mice. Bone marrow isolated from either CD4+ TCR transgenic (TCli) or CD8+ TCR transgenic (OT-1) mice was transplanted into lethally irradiated catL−/− and BL6 mice. The spleen and thymus of chimeric mice were analyzed by flow cytometry 8 wk after bone marrow transplantation. The percentage of donor T cells was determined by CD4+ Vβ6+ staining for TCli recipients (A) and CD8+ Vβ5+ staining for OT-1 recipients (B). The data shown are representative of three independent experiments with two to three mice per group.
Figure 2.
In the absence of negative selection the proportion of CD4 single-positive thymocytes is enhanced in cathepsin L–deficient mice. Bone marrow from either BL6 or Aβ−/− mice was transplanted into lethally irradiated BL6 and catL−/− mice. Mice were killed 6 wk after bone marrow transplantation and the thymi analyzed by flow cytometry to determine the percentage of CD4 single-positive thymocytes. The average percentage of CD4 single positive thymocytes in each group of chimeric animals is shown on a log scale. A total of three mice per group were analyzed in two independent experiments and each experiment is plotted separately.
Figure 3.
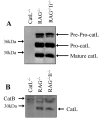
Selection of CD4+ T cells is impaired in I-Aq catL-deficient mice. (A) Thymic stromal cells were isolated from I-Ab and I-Aq catL−/−, catL+/−, and catL+/+ animals and lysed in the presence of protease inhibitors. 100 μg protein lysate was immunoprecipitated with the I-Ab,d,q specific Ab M5/114, precipitated proteins were separated by 12% SDS-PAGE and transferred to nitrocellulose before immuno-blotting with the Ii specific Ab IN-1. Binding was detected using HRP-conjugated mouse anti–rat IgG. Ii-deficient thymic stromal cells were treated as the other cell samples and used as a control for IN-1 specificity. The data is shown as different exposures of the same blot, a longer exposure was required to clearly visualize Ii fragments associated with I-Aq. The results shown are representative of two independent experiments. (B) Single cell suspensions of splenocytes and thymocytes from I-Aq catL+/− and catL−/− littermates were analyzed by flow cytometry. The CD4/CD8 profiles shown are representative of seven animals analyzed in three independent experiments.
Figure 4.
Selection of CD4+ T cells is reduced to background levels in cathepsin L x invariant chain double-deficient mice. Splenocytes (A) and thymocytes (B) were isolated from catL+/− Ii+/−, catL+/−Ii−/−, catL−/−Ii+/−, and catL−/−Ii−/− mice and analyzed by flow cytometry for CD4 and CD8. Aβ−/− mice were used as a negative control for CD4+ T cell selection. The cellularity of each thymus was ∼100 × 106. The proportion of double-positive thymocytes expressing CD69 was determined by flow cytometry (C). The data shown are representative of eight independent experiments, each with a minimum of one mouse of each genotype.
Figure 5.
Cathepsin L expression is not significantly altered in the absence of invariant chain. Thymic stromal cells were isolated from RAG−/−, RAG−/−Ii−/−, and catL−/− animals and lysed in the presence of protease inhibitors. (A) Cell lysate containing 20 μg protein was reduced, separated by SDS-PAGE in a 12% gel and the proteins electrophoretically transferred to nitrocellulose. The membrane was probed with catL specific rabbit antiserum and binding was detected using HRP-conjugated donkey anti-rabbit IgG. (B) Cell lysate containing 100 μg protein was incubated in the presence of the irreversible cysteine protease inhibitor Bio-Tyr-Ala-FMK, reduced, separated by SDS-PAGE in a 12% gel and the proteins electrophoretically transferred to nitrocellulose. Labeled proteins were detected by probing the membrane with streptavidin-conjugated HRP. These results are representative of two independent experiments.
Figure 6.
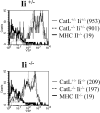
Cathepsin L plays no significant role in determining the level of expression of MHC class II on cTECs. Thymi from catL+/−Ii+/−, catL+/−Ii−/−, catL−/−Ii+/−, and catL−/−Ii−/− 6–8 week old mice were enriched for cTECs as described in the Materials and Methods. The cells were analyzed by flow cytometry and the level of expression of MHC class II on the surface of cTECs was determined by gating on CD11c−CD4−BP-1hi cells. The mean fluorescence intensity is shown in parenthesis. The data shown are representative of three independent experiments.
Similar articles
- Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus.
Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY. Nakagawa T, et al. Science. 1998 Apr 17;280(5362):450-3. doi: 10.1126/science.280.5362.450. Science. 1998. PMID: 9545226 - Expression of human cathepsin L or human cathepsin V in mouse thymus mediates positive selection of T helper cells in cathepsin L knock-out mice.
Sevenich L, Hagemann S, Stoeckle C, Tolosa E, Peters C, Reinheckel T. Sevenich L, et al. Biochimie. 2010 Nov;92(11):1674-80. doi: 10.1016/j.biochi.2010.03.014. Epub 2010 Mar 25. Biochimie. 2010. PMID: 20347002 - Proteases, processing, and thymic selection.
Cresswell P. Cresswell P. Science. 1998 Apr 17;280(5362):394-5. doi: 10.1126/science.280.5362.394. Science. 1998. PMID: 9575085 No abstract available. - The role of lysosomal proteinases in MHC class II-mediated antigen processing and presentation.
Nakagawa TY, Rudensky AY. Nakagawa TY, et al. Immunol Rev. 1999 Dec;172:121-9. doi: 10.1111/j.1600-065x.1999.tb01361.x. Immunol Rev. 1999. PMID: 10631942 Review. - Lysosomal cysteine proteases and antigen presentation.
Rudensky A, Beers C. Rudensky A, et al. Ernst Schering Res Found Workshop. 2006;(56):81-95. doi: 10.1007/3-540-37673-9_5. Ernst Schering Res Found Workshop. 2006. PMID: 16329647 Review.
Cited by
- Phagosomal proteolysis in dendritic cells is modulated by NADPH oxidase in a pH-independent manner.
Rybicka JM, Balce DR, Chaudhuri S, Allan ER, Yates RM. Rybicka JM, et al. EMBO J. 2012 Feb 15;31(4):932-44. doi: 10.1038/emboj.2011.440. Epub 2011 Dec 13. EMBO J. 2012. PMID: 22157818 Free PMC article. - Redundancy between Cysteine Cathepsins in Murine Experimental Autoimmune Encephalomyelitis.
Allan ER, Yates RM. Allan ER, et al. PLoS One. 2015 Jun 15;10(6):e0128945. doi: 10.1371/journal.pone.0128945. eCollection 2015. PLoS One. 2015. PMID: 26075905 Free PMC article. - Diversity in Cortical Thymic Epithelial Cells Occurs through Loss of a Foxn1-Dependent Gene Signature Driven by Stage-Specific Thymocyte Cross-Talk.
White AJ, Parnell SM, Handel A, Maio S, Bacon A, Cosway EJ, Lucas B, James KD, Cowan JE, Jenkinson WE, Hollander GA, Anderson G. White AJ, et al. J Immunol. 2022 Nov 14;210(1):40-9. doi: 10.4049/jimmunol.2200609. Online ahead of print. J Immunol. 2022. PMID: 36427001 Free PMC article. - Human thymoma-associated mutation of the GTF2I transcription factor impairs thymic epithelial progenitor differentiation in mice.
Giorgetti OB, Nusser A, Boehm T. Giorgetti OB, et al. Commun Biol. 2022 Sep 29;5(1):1037. doi: 10.1038/s42003-022-04002-7. Commun Biol. 2022. PMID: 36175547 Free PMC article. - Identification of Novel Molecular Markers of Human Th17 Cells.
Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M. Sałkowska A, et al. Cells. 2020 Jul 3;9(7):1611. doi: 10.3390/cells9071611. Cells. 2020. PMID: 32635226 Free PMC article.
References
- Jameson, S.C., and M.J. Bevan. 1998. T-cell selection. Curr. Opin. Immunol. 10:214–219. - PubMed
- Sant'Angelo, D.B., P.G. Waterbury, B.E. Cohen, W.D. Martin, L. Van Kaer, A.C. Hayday, and C.A. Janeway, Jr. 1997. The imprint of intrathymic self-peptides on the mature T cell receptor repertoire. Immunity. 7:517–524. - PubMed
- Barton, G.M., and A.Y. Rudensky. 1999. Requirement for diverse, low-abundance peptides in positive selection of T cells. Science. 283:67–70. - PubMed
- Villadangos, J.A., and H.L. Ploegh. 2000. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 12:233–239. - PubMed
- Riese, R.J., and H.A. Chapman. 2000. Cathepsins and compartmentalization in antigen presentation. Curr. Opin. Immunol. 12:107–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials