Inefficient enhancement of viral infectivity and CD4 downregulation by human immunodeficiency virus type 1 Nef from Japanese long-term nonprogressors - PubMed (original) (raw)
Inefficient enhancement of viral infectivity and CD4 downregulation by human immunodeficiency virus type 1 Nef from Japanese long-term nonprogressors
Minoru Tobiume et al. J Virol. 2002 Jun.
Abstract
It has been reported that patients infected with nef-defective human immunodeficiency virus type 1 (HIV-1) do not progress to AIDS; however, mutations that abrogate Nef expression are not common in long-term nonprogressors (LTNPs). We postulated that Nef function might be impaired in LTNPs, irrespective of the presence or absence of detectable amino acid sequence anomalies. To challenge this hypothesis we compared in vitro function of nef alleles that were derived from three groups of Japanese patients: LTNPs, progressors, and asymptomatic carriers (ACs). The patient-derived nef alleles were subcloned into a nef-defective infectious HIV-1 molecular clone and an expression vector. We first examined Nef-dependent enhancement of infection in a single-round infectivity assay by the use of MAGNEF cells, in which Nef is required more strictly for the infection than in the parent MAGI cells. All nef alleles from LTNPs showed reduced enhancement in the infectivity of nef-defective HIV-1 mutants compared to the nef alleles of progressors or ACs. Second, we found that nef alleles from LTNPs were less efficient in CD4 downregulation than those of progressors or ACs. Third, all nef alleles from LTNPs, progressors, and ACs reduced the cell surface expression of major histocompatibility complex class I to a similar level. Last, there was no correlation between Hck-binding activity of Nef and clinical grouping. In conclusion, we detected inefficient enhancement of HIV-1 infectivity and CD4 downregulation by HIV-1 nef alleles of LTNPs. It awaits further study to conclude that these characteristics of nef alleles are the cause or the consequence of the long-term nonprogression after HIV-1 infection.
Figures
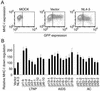
FIG. 1.
Basic vector for the analysis of Nef enhancement of infectivity. (A) The 5′ region of nef of the HIV-1 NL4-3 strain was replaced with oligonucleotides containing _Xho_I and _Not_I restriction sites to generate NL-Not. Then nef allele of NL4-3 amplified by PCR was inserted into the _Xho_I/Not_I site to generate NL-Not-Nef. (B) Wild-type NL4-3, nef_-defective NL4-3 (NL-Xh), NL-Not, and NL-Not inserted with NL4-3 nef (NL-Not-Nef) were transfected into 293T cells for propagation of virus. 293T cells were lysed and analyzed by immunoblotting with a mixture of monoclonal antibodies against p24_gag, gp41_env, and Nef. Molecular standards are shown on the left. (C) Serially diluted virus stocks were inoculated into MAGNEF cells plated on 96-well dishes. Forty-eight hours later, infected cells were identified by being stained with 5-bromo-4-chloro-3-indolyl-β-
d
-galactopyranoside. Infectivity was normalized by the quantity of p24_gag_ in the virus stocks. Experiments were done in triplicate and repeated three times. A representative result is shown with standard deviations.
FIG. 2.
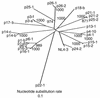
Phylogenetic analysis of the nucleotide sequences of nef alleles from LTNPs, AIDS patients (AIDS), and asymptomatic carriers (ACs). The phylogenetic tree was constructed by the neighbor-joining method. Symbols before hyphens indicate patient codes, and numbers or letters after hyphens specify each nef clone. Values along the branches indicate the bootstrap probability. All branch lengths are drawn in accordance with their relative genetic distances.
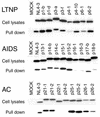
FIG. 3.
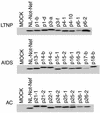
Expression of Nef from recombinant HIV-1. The sequence-verified nef alleles from LTNPs, AIDS patients, and ACs were inserted into the _Xho_I/_Not_I site of pNL-Not. The infectious DNAs were transfected into 293T cells to propagate viruses. The cell lysates were analyzed by being immunoblotted with a pool of anti-Nef monoclonal antibodies.
FIG. 4.
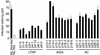
Enhancement of viral infectivity by the nef alleles derived from LTNPs, AIDS patients, or ACs. The recombinant HIV-1 viruses carrying nef alleles of patients were prepared by transfection of infectious DNAs to 293T cells. Infectivity was examined as for Fig. 1. Three independent experiments were performed, and representative data with standard deviations are shown.
FIG. 5.
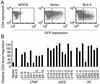
Downregulation of cell surface CD4 by Nef derived from LTNPs, AIDS patients, and ACs. (A) MAGIC5D indicator cells were transfected with pCAGGS (MOCK), pCAGGS-IRES-EGFP (Vector), or pCAGGS-IRES-EGFP-4-3-Nef (NL4-3). After 36 h, cells were stained with phycoerythrin-conjugated anti-CD4 monoclonal antibody and were analyzed for fluorescence intensity by FACSCaliber. The vertical bars indicate the thresholds for GFP. (B) pCAGGS-IRES-EGFP (Vector), pCAGGS-IRES-EGFP-4-3Nef (NL4-3), or expression vectors for the nef alleles from patients as indicated at the bottom were transfected and analyzed as described for panel A. After gating for the EGFP-positive cells, the expression level of CD4 was determined as geometric mean fluorescence with CELLQuest software. Relative downregulation efficiency (%) is shown by the following equation: (mean fluorescence intensity of cells expressing NL4-3 Nef)/(mean fluorescence intensity of cells to be evaluated) × 100. Averages from three independent experiments are shown with standard deviations.
FIG. 6.
Downregulation of cell surface MHC-I by Nef derived from LTNPs, AIDS patients, and ACs. (A) MAGIC5D indicator cells were transfected with pCAGGS (MOCK), pCAGGS-IRES-EGFP (Vector), or pCAGGS-IRES-EGFP-4-3Nef (NL4-3). After 36 h, cells were stained with phycoerythrin-conjugated anti-MHC-I monoclonal antibody and analyzed for fluorescence intensity by FACSCalibur. The vertical bars indicate the thresholds for EGFP. (B) pCAGGS-IRES-EGFP (Vector), pCAGGS-IRES-EGFP-4-3Nef (NL4-3), or expression vectors for the nef alleles from patients as indicated at the bottom were transfected and analyzed as described for panel A. After gating for EGFP-positive cells, the expression level of MHC-I was determined as geometric mean fluorescence with CELLQuest software. Relative downregulation efficiency (%) is shown by the following equation: (mean fluorescence intensity of cells expressing NL4-3 Nef)/(mean fluorescence intensity of cells to be evaluated) × 100. Averages from three independent experiments are shown with standard deviations.
FIG. 7.
Binding of Hck to the Nef proteins derived from LTNPs, AIDS patients, and ACs. 293T cells were transfected with expression plasmids encoding the nef alleles derived from patients. Two days after transfection, cells were lysed in lysis buffer, cleared by centrifugation, and incubated with GST-Hck SH2-SH3. Proteins bound to the GST fusion proteins were separated by SDS-polyacrylamide gel electrophoresis and were analyzed by being immunoblotted with a pool of anti-Nef monoclonal antibodies.
Similar articles
- Nef alleles from human immunodeficiency virus type 1-infected long-term-nonprogressor hemophiliacs with or without late disease progression are defective in enhancing virus replication and CD4 down-regulation.
Crotti A, Neri F, Corti D, Ghezzi S, Heltai S, Baur A, Poli G, Santagostino E, Vicenzi E. Crotti A, et al. J Virol. 2006 Nov;80(21):10663-74. doi: 10.1128/JVI.02621-05. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943296 Free PMC article. - CD4 and major histocompatibility complex class I downregulation by the human immunodeficiency virus type 1 nef protein in pediatric AIDS progression.
Casartelli N, Di Matteo G, Potestà M, Rossi P, Doria M. Casartelli N, et al. J Virol. 2003 Nov;77(21):11536-45. doi: 10.1128/jvi.77.21.11536-11545.2003. J Virol. 2003. PMID: 14557639 Free PMC article. - Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication.
Argañaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. Argañaraz ER, et al. J Biol Chem. 2003 Sep 5;278(36):33912-9. doi: 10.1074/jbc.M303679200. Epub 2003 Jun 19. J Biol Chem. 2003. PMID: 12816953 - Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression.
Poropatich K, Sullivan DJ Jr. Poropatich K, et al. J Gen Virol. 2011 Feb;92(Pt 2):247-68. doi: 10.1099/vir.0.027102-0. Epub 2010 Nov 24. J Gen Virol. 2011. PMID: 21106806 Review. - HIV Nef: role in pathogenesis and viral fitness.
Ariën KK, Verhasselt B. Ariën KK, et al. Curr HIV Res. 2008 May;6(3):200-8. doi: 10.2174/157016208784325001. Curr HIV Res. 2008. PMID: 18473783 Review.
Cited by
- Nef alleles from human immunodeficiency virus type 1-infected long-term-nonprogressor hemophiliacs with or without late disease progression are defective in enhancing virus replication and CD4 down-regulation.
Crotti A, Neri F, Corti D, Ghezzi S, Heltai S, Baur A, Poli G, Santagostino E, Vicenzi E. Crotti A, et al. J Virol. 2006 Nov;80(21):10663-74. doi: 10.1128/JVI.02621-05. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943296 Free PMC article. - Expression, intracellular targeting and purification of HIV Nef variants in tobacco cells.
Marusic C, Nuttall J, Buriani G, Lico C, Lombardi R, Baschieri S, Benvenuto E, Frigerio L. Marusic C, et al. BMC Biotechnol. 2007 Feb 26;7:12. doi: 10.1186/1472-6750-7-12. BMC Biotechnol. 2007. PMID: 17324250 Free PMC article. - Comprehensive analysis of nef functions selected in simian immunodeficiency virus-infected macaques.
Schindler M, Münch J, Brenner M, Stahl-Hennig C, Skowronski J, Kirchhoff F. Schindler M, et al. J Virol. 2004 Oct;78(19):10588-97. doi: 10.1128/JVI.78.19.10588-10597.2004. J Virol. 2004. PMID: 15367626 Free PMC article. - No evidence for selection of HIV-1 with enhanced gag-protease or Nef function among breakthrough infections in the CAPRISA 004 tenofovir microbicide trial.
Chopera DR, Mann JK, Mwimanzi P, Omarjee S, Kuang XT, Ndabambi N, Goodier S, Martin E, Naranbhai V, Karim SA, Karim QA, Brumme ZL, Ndung'u T, Williamson C, Brockman MA; CAPRISA 004 TRAPS Team. Chopera DR, et al. PLoS One. 2013 Aug 28;8(8):e71758. doi: 10.1371/journal.pone.0071758. eCollection 2013. PLoS One. 2013. PMID: 24015191 Free PMC article.
References
- Adachi, A., N. Ono, H. Sakai, K. Ogawa, R. Shibata, T. Kiyomasu, H. Masuike, and S. Ueda. 1991. Generation and characterization of the human immunodeficiency virus type 1 mutants. Arch. Virol. 117:45-58. - PubMed
- Buchbinder, S., and E. Vittinghoff. 1999. HIV-infected long-term nonprogressors: epidemiology, mechanisms of delayed progression, and clinical and research implications. Microbes Infect. 1:1113-1120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials