Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate - PubMed (original) (raw)
. 2002 Jun;76(12):6277-92.
doi: 10.1128/jvi.76.12.6277-6292.2002.
Joann Taylor, Geoffrey H Holm, Andrew Mehle, Tom Morgan, Mark Cayabyab, Michael Farzan, Hui Wang, Jeanne E Bell, Kevin Kunstman, John P Moore, Steven M Wolinsky, Dana Gabuzda
Affiliations
- PMID: 12021361
- PMCID: PMC136234
- DOI: 10.1128/jvi.76.12.6277-6292.2002
Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate
Paul R Gorry et al. J Virol. 2002 Jun.
Abstract
Most human immunodeficiency virus type 1 (HIV-1) viruses in the brain use CCR5 as the principal coreceptor for entry into a cell. However, additional phenotypic characteristics are necessary for HIV-1 neurotropism. Furthermore, neurotropic strains are not necessarily neurovirulent. To better understand the determinants of HIV-1 neurovirulence, we isolated viruses from brain tissue samples from three AIDS patients with dementia and HIV-1 encephalitis and analyzed their ability to induce syncytia in monocyte-derived macrophages (MDM) and neuronal apoptosis in primary brain cultures. Two R5X4 viruses (MACS1-br and MACS1-spln) were highly fusogenic in MDM and induced neuronal apoptosis. The R5 viruses UK1-br and MACS2-br are both neurotropic. However, only UK1-br induced high levels of fusion in MDM and neuronal apoptosis. Full-length Env clones from UK1-br required lower CCR5 and CD4 levels than Env clones from MACS2-br to function efficiently in cell-to-cell fusion and single-round infection assays. UK1-br Envs also had a greater affinity for CCR5 than MACS2-br Envs in binding assays. Relatively high levels of UK1-br and MACS2-br Envs bound to CCR5 in the absence of soluble CD4. However, these Envs could not mediate CD4-independent infection, and MACS2-br Envs were unable to mediate fusion or infection in cells expressing low levels of CD4. The UK1-br virus was more resistant than MACS2-br to inhibition by the CCR5-targeted inhibitors TAK-779 and Sch-C. UK1-br was more sensitive than MACS2-br to neutralization by monoclonal antibodies (2F5 and immunoglobulin G1b12 [IgG1b12]) and CD4-IgG2. These results predict the presence of HIV-1 variants with increased CCR5 affinity and reduced dependence on CCR5 and CD4 in the brains of some AIDS patients with central nervous system disease and suggest that R5 variants with increased CCR5 affinity may represent a pathogenic viral phenotype contributing to the neurodegenerative manifestations of AIDS.
Figures
FIG. 1.
Syncytium formation in MDM induced by neurotropic HIV-1 isolates. (A) MDM were infected with equivalent amounts of HIV-1 as described in Materials and Methods. Syncytia formation was observed at day 7 (ADA, MACS1-spln, UK1-br, MACS2-br) or 10 (MACS1-br) postinfection. Control cells were exposed to the X4 primary virus isolate CB1-br, which does not replicate in MDM (32). (B) Multinucleated giant cells in brain tissue sections of patient UK1 stained with hematoxylin and eosin (left panel) and immunocytochemical detection of HIV-1 p24 antigen in brain tissue (right panel). Photographs were taken using an Eclipse TE 300 microscope (Nikon, Osaka, Japan) at a magnification of ×400.
FIG. 2.
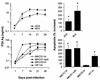
Apoptosis induced by infection of primary brain cultures with neurotropic HIV-1 isolates. Control viruses (ADA, 89.6) or primary virus isolates (MACS1-br, MACS1-spln, MACS2-br, UK1-br) were used at equivalent amounts to infect microglia in mixed brain cell cultures. Virus replication was measured weekly by HIV-1 p24 production in culture supernatants for 28 days postinfection. Apoptosis of neurons was measured at day 28 as described in Materials and Methods. Data are expressed as the percent change in comparison to control cells treated with culture medium alone, with the control value set at 0%. Data shown are means of duplicate infections, with error bars representing standard deviations. Results are representative of three independent experiments with cells obtained from different donors. Asterisk, P < 0.05 for the difference in apoptosis levels in a comparison with mock-infected cultures by Student's t test.
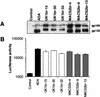
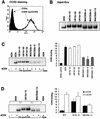
FIG. 3.
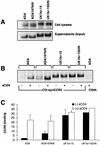
Expression and cell-to-cell fusion activity of full-length Env clones. Gp160 Env genes of virus isolates UK1-br and MACS2-br were cloned into the pCR3.1 expression vector (Invitrogen) from genomic DNA of PBMC infected with each isolate, as described in Materials and Methods. 293T cells were cotransfected with 10 μg (each) of Env plasmid plus 2 μg of pLTR-Tat. (A) Env expression at 72 h posttransfection was measured by Western blot analysis of cell lysates using rabbit anti-gp120 polyclonal antisera. Positions of gp160 and gp120 are indicated on the right. (B) Fusion assays were performed using 293T cells expressing each Env and Cf2-Luc cells expressing CD4 and CCR5 as described in Materials and Methods. Data are expressed as means from duplicate experiments. Error bars represent standard deviations.
FIG. 4.
Env amino acid sequences. Full-length HIV-1 gp160 amino acid sequences were obtained from Env genes cloned into pCR3.1 as described in Materials and Methods. Amino acid alignments are compared to Env from HIV-1 ADA and the clade B consensus sequence. Dots indicate residues identical to the clade B consensus, and dashes indicate gaps. Note the asparagine-to-aspartic acid substitution at position 208 and the 12-amino-acid insertion in V1 for UK1-br Env clones.
FIG. 4.
Env amino acid sequences. Full-length HIV-1 gp160 amino acid sequences were obtained from Env genes cloned into pCR3.1 as described in Materials and Methods. Amino acid alignments are compared to Env from HIV-1 ADA and the clade B consensus sequence. Dots indicate residues identical to the clade B consensus, and dashes indicate gaps. Note the asparagine-to-aspartic acid substitution at position 208 and the 12-amino-acid insertion in V1 for UK1-br Env clones.
FIG. 5.
Effect of CD4 and CCR5 levels on cell-to-cell fusion and HIV-1 entry. Cf2th cells were transfected with 0.05, 0.5, 5, or 10 μg of CD4- and CCR5-expressing plasmid and analyzed for surface expression of CD4 or CCR5 by flow cytometry (A). Similar results were obtained with transfected Cf2-Luc cells (data not shown). Cf2-Luc (B) or Cf2th (C) cells were cotransfected with 0.05, 0.5, 5, or 10 μg of CD4-expressing plasmid and 0.05, 0.5, 5, or 10 μg of CCR5-expressing plasmid to generate 16 populations of cells expressing different combinations of each receptor. The total amount of DNA in each transfection was adjusted to 20 μg with pcDNA3. 293T cells cotransfected with 10 μg of Env plasmid and 2 μg of pLTR-Tat were used in fusion assays with transfected Cf2-Luc cells as described in Materials and Methods (B). Luciferase reporter viruses pseudotyped with each Env were used to infect transfected Cf2th cells as described in Materials and Methods (C). Data are expressed as means from duplicate experiments. Error bars represent standard deviations. Similar results were obtained in two independent experiments.
FIG. 6.
CCR5 binding assays. Cf2th-synCCR5 cells were analyzed for surface expression of CCR5 by flow cytometry (A). Equivalent amounts of radiolabeled soluble Env proteins (B) were used in binding assays with Cf2-synCCR5 cells in the presence of sCD4 (C) or in the presence and absence of sCD4 (D). Cells were washed and lysed, and the bound Env glycoproteins were immunoprecipitated using AIDS patient serum and protein A Sepharose beads. Proteins were analyzed using SDS-PAGE (C and D, left) and then quantitated by densitometry (C and D, right). The gels shown in panels C and D are representative of those from two independent experiments. The quantitation of CCR5 binding shown in panels C and D represent means and standard deviations of values obtained in two independent experiments, normalized against background levels of ADA Env bound to the parental Cf2th cell line.
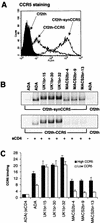
FIG. 7.
CCR5 binding assays with cells expressing different amounts of CCR5. Cf2th-synCCR5 and Cf2th-CCR5 cells were analyzed for surface expression of CCR5 by flow cytometry (A). Equivalent amounts of radiolabeled soluble Env proteins (see Fig. 6A) were used in binding assays with Cf2th-synCCR5 or Cf2th-CCR5 cells in the presence of sCD4. Cells were washed and lysed, and the bound Env glycoproteins were immunoprecipitated using AIDS patient serum and protein A Sepharose beads. Proteins were analyzed using SDS-PAGE (B) and then quantitated by densitometry (C). The gels shown in panel B are representative of those from two independent experiments. The quantitation of CCR5 binding shown in panel C represents means and standard deviations of values obtained in two independent experiments, normalized against background levels of ADA Env bound to the parental Cf2th cell line.
FIG. 8.
Effect of N-linked glycosylation on CCR5 binding and CD4 dependence. Env glycoproteins were immunoprecipitated from cell lysates or supernatants of radiolabeled 293T cells transfected with wild-type ADA or UK1-br Env plasmids or with mutants containing a loss of a potential glycosylation site at position 197 (ADA197N/K) or a restored glycosylation site at position 208 (UK1br-15D/N) (A). Equivalent amounts of radiolabeled soluble Env proteins were used in binding assays with Cf2th-synCCR5 cells in the presence or absence of sCD4. Cells were washed and lysed, and the bound Env glycoproteins were immunoprecipitated using AIDS patient serum and protein A Sepharose beads. Proteins were analyzed using SDS-PAGE (B) and then quantitated by densitometry (C). The gel shown in panel B is representative of two independent experiments. The quantitation of CCR5 binding shown in panel C represents means and standard deviations of values obtained in two independent experiments, normalized against background levels of ADA Env bound to the parental Cf2th cell line.
FIG. 9.
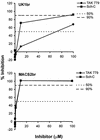
Sensitivity of UK1-br and MACS2-br viruses to CCR5 inhibitors. PBMC were treated with the CCR5 antagonists TAK-779 and Sch-C at concentrations increasing by 10-fold from 0.01 to 100 μM and then infected with the UK1-br and MACS2-br primary isolates. Virus production postinfection was assessed at day 14 by measuring the amount of soluble HIV-1 p24 antigen in the culture supernatant (82). The production of p24 antigen in inhibitor-treated cultures was calculated as a percentage of that produced in the absence of an inhibitor (defined as 100%). The mean values obtained from an assay performed in duplicate are shown and are representative of those from two independent experiments. Similar results were obtained at days 7 and 10 postinfection (data not shown).
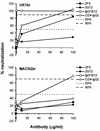
FIG. 10.
Sensitivity of UK1-br and MACS2-br viruses to neutralization. Each virus was incubated for 30 min with MAbs or CD4-IgG2 at concentrations increasing by 10-fold from 0.01 to 100 μg/ml. The mixtures were then added to PBMC. Virus production in culture supernatants at day 14 postinfection was measured by quantitation of soluble HIV-1 p24 antigen (82). Virus replication and its inhibition were measured and calculated as described in the legend to Fig. 9. The mean values obtained from an assay performed in duplicate are shown.
Similar articles
- HIV-1 R5 Macrophage-Tropic Envelope Glycoprotein Trimers Bind CD4 with High Affinity, while the CD4 Binding Site on Non-macrophage-tropic, T-Tropic R5 Envelopes Is Occluded.
Quitadamo B, Peters PJ, Repik A, O'Connell O, Mou Z, Koch M, Somasundaran M, Brody R, Luzuriaga K, Wallace A, Wang S, Lu S, McCauley S, Luban J, Duenas-Decamp M, Gonzalez-Perez MP, Clapham PR. Quitadamo B, et al. J Virol. 2018 Jan 2;92(2):e00841-17. doi: 10.1128/JVI.00841-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118121 Free PMC article. - Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS.
Sterjovski J, Churchill MJ, Ellett A, Gray LR, Roche MJ, Dunfee RL, Purcell DF, Saksena N, Wang B, Sonza S, Wesselingh SL, Karlsson I, Fenyo EM, Gabuzda D, Cunningham AL, Gorry PR. Sterjovski J, et al. Retrovirology. 2007 Dec 12;4:89. doi: 10.1186/1742-4690-4-89. Retrovirology. 2007. PMID: 18076768 Free PMC article. - Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion.
Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. Thomas ER, et al. Virology. 2007 Mar 30;360(1):105-19. doi: 10.1016/j.virol.2006.09.036. Epub 2006 Nov 7. Virology. 2007. PMID: 17084877 Free PMC article. - Variation of macrophage tropism among HIV-1 R5 envelopes in brain and other tissues.
Peters PJ, Dueñas-Decamp MJ, Sullivan WM, Clapham PR. Peters PJ, et al. J Neuroimmune Pharmacol. 2007 Mar;2(1):32-41. doi: 10.1007/s11481-006-9042-2. Epub 2006 Nov 7. J Neuroimmune Pharmacol. 2007. PMID: 18040824 Review. - Mechanisms of HIV-1 neurotropism.
Dunfee R, Thomas ER, Gorry PR, Wang J, Ancuta P, Gabuzda D. Dunfee R, et al. Curr HIV Res. 2006 Jul;4(3):267-78. doi: 10.2174/157016206777709500. Curr HIV Res. 2006. PMID: 16842080 Review.
Cited by
- Changes in cerebrospinal fluid proteins across the spectrum of untreated and treated chronic HIV-1 infection.
Hu Z, Cinque P, Dravid A, Hagberg L, Yilmaz A, Zetterberg H, Fuchs D, Gostner J, Blennow K, Spudich SS, Kincer L, Zhou S, Joseph SB, Swanstrom R, Price RW, Gisslén M. Hu Z, et al. PLoS Pathog. 2024 Sep 24;20(9):e1012470. doi: 10.1371/journal.ppat.1012470. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39316609 Free PMC article. - When human immunodeficiency virus meets chemokines and microglia: neuroprotection or neurodegeneration?
Mocchetti I, Campbell LA, Harry GJ, Avdoshina V. Mocchetti I, et al. J Neuroimmune Pharmacol. 2013 Mar;8(1):118-31. doi: 10.1007/s11481-012-9353-4. Epub 2012 Apr 15. J Neuroimmune Pharmacol. 2013. PMID: 22527632 Free PMC article. Review. - Pathogenicity and immunogenicity of attenuated, nef-deleted HIV-1 strains in vivo.
Gorry PR, McPhee DA, Verity E, Dyer WB, Wesselingh SL, Learmont J, Sullivan JS, Roche M, Zaunders JJ, Gabuzda D, Crowe SM, Mills J, Lewin SR, Brew BJ, Cunningham AL, Churchill MJ. Gorry PR, et al. Retrovirology. 2007 Sep 23;4:66. doi: 10.1186/1742-4690-4-66. Retrovirology. 2007. PMID: 17888184 Free PMC article. - Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss.
Anastassopoulou CG, Marozsan AJ, Matet A, Snyder AD, Arts EJ, Kuhmann SE, Moore JP. Anastassopoulou CG, et al. PLoS Pathog. 2007 Jun;3(6):e79. doi: 10.1371/journal.ppat.0030079. PLoS Pathog. 2007. PMID: 17542646 Free PMC article. - HIV evolution and escape.
Richman DD, Little SJ, Smith DM, Wrin T, Petropoulos C, Wong JK. Richman DD, et al. Trans Am Clin Climatol Assoc. 2004;115:289-303. Trans Am Clin Climatol Assoc. 2004. PMID: 17060974 Free PMC article. Review.
References
- Adie-Biassette, H., Y. Levy, M. Colombel, F. Poron, S. Natchev, C. Keohane, and F. Gray. 1995. Neuronal apoptosis in HIV infection in adults. Neuropathol. Appl. Neurobiol. 21:218-227. - PubMed
- Albright, A. V., J. T. C. Shieh, T. Itoh, B. Lee, D. Pleasure, M. J. O'Connor, R. W. Doms, and F. Gonzalez-Scarano. 1999. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J. Virol. 73:205-213. - PMC - PubMed
- Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. - PubMed
- Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M.-C. Gauduin, R. A. Koup, S. J. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4IgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res. Hum. Retrovir. 11:533-540. - PubMed
- An, S. F., F. Gray, and F. Scaravalli. 1995. Programmed cell death in brains of HIV-1-positive pre-AIDS patients. Lancet 346:911-912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS35734/NS/NINDS NIH HHS/United States
- P30 AI028691/AI/NIAID NIH HHS/United States
- R01 NS035734/NS/NINDS NIH HHS/United States
- AI41420/AI/NIAID NIH HHS/United States
- CA79458/CA/NCI NIH HHS/United States
- R01 NS037277/NS/NINDS NIH HHS/United States
- DA13127/DA/NIDA NIH HHS/United States
- NS37277/NS/NINDS NIH HHS/United States
- AI28691/AI/NIAID NIH HHS/United States
- R01 AI041420/AI/NIAID NIH HHS/United States
- U01 AI35039/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous