The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription - PubMed (original) (raw)
The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription
Suzanne D Turner et al. Mol Cell Biol. 2002 Jun.
Abstract
Transcription of the Saccharomyces cerevisiae ARG1 gene is under the control of both positive and negative elements. Activation of the gene in minimal medium is induced by Gcn4. Repression occurs in the presence of arginine and requires the ArgR/Mcm1 complex that binds to two upstream arginine control (ARC) elements. With the recent finding that the E2 ubiquitin conjugase Rad6 modifies histone H2B, we examined the role of Rad6 in the regulation of ARG1 transcription. We find that Rad6 is required for repression of ARG1 in rich medium, with expression increased approximately 10-fold in a rad6 null background. Chromatin immunoprecipitation analysis indicates increased binding of TATA-binding protein in the absence of Rad6. The active-site cysteine of Rad6 is required for repression, implicating ubiquitination in the process. The effects of Rad6 at ARG1 involve two components. In one of these, histone H2B is the likely target for ubiquitination by Rad6, since a strain expressing histone H2B with the principal ubiquitination site converted from lysine to arginine shows a fivefold relief of repression. The second component requires Ubr1 and thus likely the pathway of N-end rule degradation. Through the analysis of promoter constructs with ARC deleted and an arg80 rad6 double mutant, we show that Rad6 repression is mediated through the ArgR/Mcm1 complex. In addition, analysis of an ada2 rad6 deletion strain indicated that the SAGA acetyltransferase complex and Rad6 act in the same pathway to repress ARG1 in rich medium.
Figures
FIG. 1.
Rad6 is required for the repression the ARG1 promoter in rich medium. (A) _ARG1_-lacZ reporter construct. ARG1 sequences from −640 to +213 were cloned as a _Bam_HI-_Hin_dIII fragment into YCp87 to generate a _his3_-lacZ translational fusion on a LEU2 centromeric plasmid. Previously mapped regulatory sites (19) are shown as follows: ARC elements (centered at −175 and −214 relative to the principal transcriptional start site [16]), A TATA elements at −73 is not shown. Gcn4 binding sites (−195 and −265), and the Abf1 consensus sequence (−302). (B) β-Galactosidase analysis of _ARG1_-lacZ expression in wild-type (WT) and rad6 deletion backgrounds. Yeast strains BY4741 (wild type) and BY4425 (rad6_Δ_0) containing YCp87-_ARG1_-lacZ were grown to saturation in minimal medium and then diluted 1/100 in YPD medium or minimal medium (2% glucose supplemented with the required amino acids). The cells were harvested at an _A_600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean for two experiments performed in triplicate.
FIG. 2.
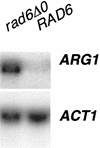
Northern analysis of ARG1 in a rad6 disruption strain. Yeast strains BY4741 (wild type) and BY4425 (rad6_Δ_0) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot shown is representative of analyses performed in triplicate.
FIG. 3.
Rad6 inhibits the binding of TBP to the ARG1 promoter. BY4741 (wild type [WT]) and BY4425 (rad6_Δ_0) containing HA epitope-tagged TBP were grown in 150 ml of YPD medium and cross-linked with 1% formaldehyde followed by mock immunoprecipitation with no antibody (−; lanes 1 and 2) or immunoprecipitation with anti-HA (α-HA) antibody (+; lanes 3 and 4). Immunoprecipitated DNA and input DNA (lanes 5 and 6) were analyzed by PCR using primers specific for ARG1 and the ACT1 promoter. Linear ranges for PCR were determined by serial dilution. The purified extended products were analyzed on 6% native polyacrylamide gels and stained with ethidium bromide. The data are representative of three independent whole-cell extracts and ChIP assays.
FIG. 4.
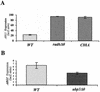
(A) Ubiquitin conjugase activity of Rad6 is required for its repression of ARG1 expression. BY4425 (rad6_Δ_0) containing YCp87-_ARG1_-lacZ and either no plasmid (rad6_Δ_0), a HIS3 centromeric plasmid expressing wild-type Rad6 (HH1; WT), or Rad6-C88A (HH4; C88A) (35) was grown to saturation in minimal medium and then diluted 1/100 in YPD medium. The cells were harvested at an _A_600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean of three samples. (B) Deletion of UBP3 results in reduced expression of ARG1 in rich medium. Yeast strains BY4741 and BY16148 (ubp3_Δ_0) containing _ARG1_-lacZ were grown, and β-galactosidase activity was determined as described above (n = 4; P = 0.04). The error bars represent the standard error of the mean.
FIG. 5.
A K123R mutation within histone H2B results in relief of repression of the ARG1 promoter. (A) An allele of HTB1 containing a K123R mutation at the site of ubiquitination was introduced by gene replacement as the only cellular copy of histone H2B (yeast strain CY1272; H2BK-R). The isogenic strain CY1256 (WT) expressing wild-type H2B was similarly constructed. Yeast strain CY1284 (H2BK-R rad6_Δ_0) was constructed by disrupting rad6 in the CY1272 background. These strains, as well as BY4425 (rad6_Δ_0), all containing YCp87-_ARG1_-lacZ, were grown to saturation in minimal medium and then diluted 1/100 in YPD medium. The cells were harvested at an _A_600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean of four samples. (B) Northern analysis. Yeast strains BY4425 (rad6_Δ_0), CY1256 (htb2_Δ_0), CY1272 (H2BK-R htb2_Δ_0), and BY4741 (wild type) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot is representative of analyses performed in triplicate. Two independent RNA preparations from CY1272 are shown.
FIG. 6.
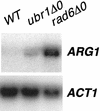
ARG1 expression in a UBR1 deletion strain . Yeast strains BY4741 (wild type [WT]), BY4425 (rad6_Δ_0), and BY14814 (ubr1_Δ_0) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot shown is representative of three experiments.
FIG. 7.
Recruitment of Rad6 to the promoter requires components of the ArgR/Mcm1 complex. Saturated cultures of BY4425 (rad6_Δ_0), BY14814 (ubr1_Δ_0), and BY4741 (wild type [WT]) containing YCp87-_ARG1_-lacZ, YCp87-_ARG1_ΔABF1-lacZ, or YCp87-_ARG1_ΔARC-lacZ were grown in minimal medium and then diluted 1/100 in YPD. The cultures were grown to an _A_600 of 1.0 to 1.5. β-Galactosidase activity was determined using ONPG as the substrate, standardizing to cell density. lacZ units represent the averages of the mean for an experiment performed in triplicate with a standard error of the mean under 10%. The ratio of lacZ expression in the null strain versus the wild-type (wt) strain is shown for each lacZ fusion construct. nd, not determined.
Similar articles
- Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium.
Ricci AR, Genereaux J, Brandl CJ. Ricci AR, et al. Mol Cell Biol. 2002 Jun;22(12):4033-42. doi: 10.1128/MCB.22.12.4033-4042.2002. Mol Cell Biol. 2002. PMID: 12024017 Free PMC article. - Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae.
Crabeel M, de Rijcke M, Seneca S, Heimberg H, Pfeiffer I, Matisova A. Crabeel M, et al. Yeast. 1995 Nov;11(14):1367-80. doi: 10.1002/yea.320111405. Yeast. 1995. PMID: 8585320 - Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter.
Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Wood A, et al. Mol Cell. 2003 Jan;11(1):267-74. doi: 10.1016/s1097-2765(02)00802-x. Mol Cell. 2003. PMID: 12535539 - The structure and function of RAD6 and RAD18 DNA repair genes of Saccharomyces cerevisiae.
Prakash L. Prakash L. Genome. 1989;31(2):597-600. doi: 10.1139/g89-111. Genome. 1989. PMID: 2698834 Review. - The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond.
Deng ZH, Ai HS, Lu CP, Li JB. Deng ZH, et al. Chromosome Res. 2020 Dec;28(3-4):247-258. doi: 10.1007/s10577-020-09640-3. Epub 2020 Sep 7. Chromosome Res. 2020. PMID: 32895784 Review.
Cited by
- Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10.
Carvin CD, Kladde MP. Carvin CD, et al. J Biol Chem. 2004 Aug 6;279(32):33057-62. doi: 10.1074/jbc.M405033200. Epub 2004 Jun 4. J Biol Chem. 2004. PMID: 15180994 Free PMC article. - Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability.
Chandrasekharan MB, Huang F, Sun ZW. Chandrasekharan MB, et al. Proc Natl Acad Sci U S A. 2009 Sep 29;106(39):16686-91. doi: 10.1073/pnas.0907862106. Epub 2009 Sep 10. Proc Natl Acad Sci U S A. 2009. PMID: 19805358 Free PMC article. - Potent macrocycle inhibitors of the human SAGA deubiquitinating module.
Morgan M, Ikenoue T, Suga H, Wolberger C. Morgan M, et al. Cell Chem Biol. 2022 Apr 21;29(4):544-554.e4. doi: 10.1016/j.chembiol.2021.12.004. Epub 2021 Dec 21. Cell Chem Biol. 2022. PMID: 34936860 Free PMC article. - The Paf1 complex represses ARG1 transcription in Saccharomyces cerevisiae by promoting histone modifications.
Crisucci EM, Arndt KM. Crisucci EM, et al. Eukaryot Cell. 2011 Jun;10(6):712-23. doi: 10.1128/EC.05013-11. Epub 2011 Apr 15. Eukaryot Cell. 2011. PMID: 21498644 Free PMC article. - The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination.
Long L, Thelen JP, Furgason M, Haj-Yahya M, Brik A, Cheng D, Peng J, Yao T. Long L, et al. J Biol Chem. 2014 Mar 28;289(13):8916-30. doi: 10.1074/jbc.M114.551754. Epub 2014 Feb 13. J Biol Chem. 2014. PMID: 24526689 Free PMC article.
References
- Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous