Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants - PubMed (original) (raw)
Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants
Eleni Goshu et al. Mol Cell Biol. 2002 Jun.
Abstract
The mouse genome contains two Sim genes, Sim1 and Sim2. They are presumed to be important for central nervous system (CNS) development because they are homologous to the Drosophila single-minded (sim) gene, mutations in which cause a complete loss of CNS midline cells. In the mammalian CNS, Sim2 and Sim1 are coexpressed in the paraventricular nucleus (PVN). While Sim1 is essential for the development of the PVN (J. L. Michaud, T. Rosenquist, N. R. May, and C.-M. Fan, Genes Dev. 12:3264-3275, 1998), we report here that Sim2 mutant has a normal PVN. Analyses of the Sim1 and Sim2 compound mutants did not reveal obvious genetic interaction between them in PVN histogenesis. However, Sim2 mutant mice die within 3 days of birth due to lung atelectasis and breathing failure. We attribute the diminished efficacy of lung inflation to the compromised structural components surrounding the pleural cavity, which include rib protrusions, abnormal intercostal muscle attachments, diaphragm hypoplasia, and pleural mesothelium tearing. Although each of these structures is minimally affected, we propose that their combined effects lead to the mechanical failure of lung inflation and death. Sim2 mutants also develop congenital scoliosis, reflected by the unequal sizes of the left and right vertebrae and ribs. The temporal and spatial expression patterns of Sim2 in these skeletal elements suggest that Sim2 regulates their growth and/or integrity.
Figures
FIG. 1.
Generation of the Sim2 mutant. (A) The top line shows the genomic organization of the wild-type (wt) Sim2 locus, the middle line shows the homologous recombination vector (HR), and the bottom line shows the recombined mutant allele (m). PGK-neo and PGK-tk are cassettes for positive and negative selections. _Bam_HI, B; _Eco_RI, R; _Not_I, N. The size bar = 1 kb. Arrowheads indicate the positions of the primers used for PCR, and shaded boxes indicate the 5′ and 3′ probes. (B) Southern blot analyses with the 5′ (left) and 3′ (right) probes in panel A. _Eco_RI was used to digest the genomic DNA of the wild type (+/+), heterozygous (+/−), and mutant (−/−) mice. The size markers (kilobases) and the wild-type and mutant bands (in kilobases) are as labeled. (C) PCR of the genomic DNA to show that Sim2 bHLH region is deleted in the mutant. The neo sequence is detected as a 189-bp fragment, while the Sim2 bHLH domain is detected as a 135-bp fragment. Con, without DNA input; M, 123-bp marker. (D) Section in situ hybridization of wild-type and mutant kidneys with a 35S-UTP-labeled Sim2 3′UTR (not deleted in the mutant) probe. The top panels show the phase images, and the bottom panels show the corresponding dark-field images. Sim2 is normally expressed in the collecting ducts, but not in the mutant (black and white arrowheads).
FIG. 2.
Expression pattern of Sim2. In situ hybridization with 35S-labeled Sim2 probe was used to assess the Sim2 expression pattern. Embryos were Carnoy's solution fixed, paraffin embedded, and sectioned at 8 μm. The histology of the sections is on the left next to the corresponding dark-field image revealing Sim2 expression (the silver granules). (A and A′) Midsagittal section of an E9.5 embryo. di, diencephalon. (B and B′) Transverse sections of an E10.5 embryo. (C and C′) Sagittal sections of an E10.5 embryo. sc, sclerotome; br, branchial arches; h, heart; lb, limb. (D and D′) Transverse sections of an E11.5 embryo. (E and E′) Sagittal sections of an E11.5 embryo. ar, vertebral arch; vb, vertebral body; r, ribs; L, lung; pw, pleural wall. (F and F′) Transverse sections of an E12.5 embryo. (G and G′) Sagittal sections of an E12.5 embryo. (L) Sagittal section of an E16.5 embryo. Sim2 shows expression in the PVN, kidney (k), muscles in the genital tubercles (gt), palate (p), nasal pit (np), tongue (t), ribs (r), vertebra (v), diaphragm (dia), and trachea (tr). st, sternum; H, heart. Higher magnifications of sagittal sections of E16.5 pleural cavity (H and H′) and diaphragm (K and K′) and transverse sections of vertebra and erector muscles (I and I′) and ribs (J and J′) are also shown. Arrowheads indicate _Sim2-_positive structures.
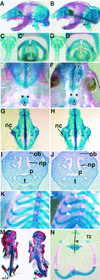
FIG.3.
Axial skeleton phenotype of the Sim2 mutant: Shown are skeleton preparations of the newborn wild-type (+/+) (A, C, C′, E, G, I, and K) and mutant Sim2 (−/−) (B, D, D′, F, H, J, and L) siblings with alcian blue and alizarin red. Panels A and B show sagittal views of the heads. Panels C and D show frontal views of the trachea. Panels C′ and D′ show top views of the trachea and epiglottis (open arrowheads). Panels E and F show ventral views of the palate. Asterisks label the palatal bones, and arrowheads indicate the fusion midlines. Panels G and H show dorsal views of the isolated nasal cartilage (nc). Panels I and J show coronal sections of the heads stained with hematoxylin. ob, olfactory bulb; p, palate; t, tongue; np, nasal pit. Panels K and L show frontal partial views of the rib cages. White arrowheads indicate the outgrowths on the ribs in panel L. Panel M shows skeleton preparation illustrating the scoliosis of the Sim2 mutant on the right (−/−); the wild-type littermate is on the left (+/+). Panel N shows an example of an asymmetric T2 vertebra/rib element from a mutant. The bent line through the center of the vertebral body and the spinal process indicate that the left arch is larger, a cause of scoliosis.
FIG. 4.
Multiple phenotypes of the Sim2 mutant may contribute to lung atelectasis. Panels A and B show two examples of 12-h BrdU-labeled E18.5 ribs. Proliferating cells (HRP-positive brown nuclei) were found in the rib protrusions. r, rib; s, sternum. Black lines outline the protrusions. Panels C and D show two examples of the PAS-stained rib protrusions (open arrowheads) and their connections to the intercostal muscle. A solid arrowhead marks the extension off of some protrusions. Panels E (+/+) and E′ (−/−) show Mallory's staining of the pleural mesothelium basement membrane in the mutant indicating it was disrupted after the mutant had shown severe dyspnea. Before severe dyspnea, however, the mutant mesothelium appeared normal (data not shown). Higher magnification of the mesothelium (arrowhead) is to the right of each figure in black frames. Panels F (+/+) and F′ (−/−) show 2H3 (antineurofilament) staining of the innervated nerves (arrowheads) in the diaphragm (black brackets). Panels G (+/+) and G′ (−/−) show MF20 (antimyosin) staining of muscle fibers in the diaphragm (white brackets). Table 2 provides a summary of the frequency and phenotype of rib protrusions and scoliosis.
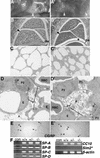
FIG. 5.
Lung phenotype of the Sim2 mutant. Panels A (+/+) and A′ (−/−) show whole-mount live lungs in the chest cavities of newborn wild-type and Sim2 mutant mice. Panels B (+/+) and B′ (−/−) show sagittal sections of newborn lungs stained with hematoxylin and eosin. dia, diaphragm. Asterisks indicate the space between the lung and the diaphragm in the mutant. Arrowheads indicate the part of the lung that is the most different in histology. Panels C (+/+) and C′ (−/−) are 1-μm plastic sections of the lungs at magnifications of ×100. Atelectasis in the mutant is evident by the smaller alveolar opening and thicker alveolar wall. Panels D (+/+) and D′ (−/−) show electron microscopy of the alveolus (A), endothelium (E), capillary, erythrocyte (RBC), type I (P1) and II (P2) pneumocytes, lamina body (L), and the surfactant (×1,650). Atmagnifications of ×2,100 and ×4,400, intact basement membranes, cell junctions, smooth muscle actins, and lamina body structures were seen in the mutant. In other fields, macrophages and fibroblast supporting cells also were normal in their position and integrity (data not shown). Panels E (+/+) and E′ (−/−) show CGRP staining of the neuroendocrine cells in the airway epithelium (arrowheads). (F) RT-PCR assay for expression of SP-A, -B, -C, and -D and CC-10, (30 cycles), SIM2 (35 cycles), and β-actin (25 cycles) in the lung (see Materials and Methods). The primers for Sim2 could detect the correct size of product in the kidney RNA samples with 30 cycles of PCR (not shown). β-Actin was assayed for normalization. con, no RNA control.
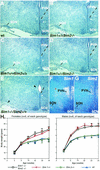
FIG. 6.
Sim2 has no obvious genetic interaction with Sim1 in PVN development and energy homeostasis. Brains of wild-type (Sim1+/+/Sim2+/+) (A), Sim2 mutant (Sim1+/+/_Sim2_−/−) (B), Sim1 heterozygous (Sim1 +/−/Sim2+/+) (C), Sim1 heterozygote/Sim2 mutant (Sim1+/−/_Sim2_−/−) (D), and Sim1 mutant (_Sim1_−/−) (E) mice were embedded in paraffin, sectioned, and stained with hematoxylin. SCN, suprachiasmatic nucleus. An asterisk labels the medial preoptic nucleus as a reference. Dashed lines outline the ventral boundary of the PVN. Arrowheads in panel E indicate the lack of PVN and SON in _Sim1_−/− mice. (F and G) 35S radioactive in situ hybridization of the PVN/SON region with Sim1 and Sim2 probes. (H) Six females (left) and five males (right) of each of the following genotypes were used in this study: wild type (wt, triangles), Sim1+/− (diamonds), Sim2+/− (circles), and Sim1+/−/Sim2+/− (squares). Animals were weighed every 4 weeks after birth, continuing on until 30 weeks. The average weight (in grams) was plotted, and the error bars represent standard errors. By Student's t test, there is no significant difference between wild-type and Sim2 heterozygous mice (P > 0.75) or between Sim1+/− and Sim2+/−/Sim1+/− mice (P > 0.8).
Similar articles
- Sim1 and Sim2 expression during chick and mouse limb development.
Coumailleau P, Duprez D. Coumailleau P, et al. Int J Dev Biol. 2009;53(1):149-57. doi: 10.1387/ijdb.082659pc. Int J Dev Biol. 2009. PMID: 19123137 - Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus.
Goshu E, Jin H, Lovejoy J, Marion JF, Michaud JL, Fan CM. Goshu E, et al. Mol Endocrinol. 2004 May;18(5):1251-62. doi: 10.1210/me.2003-0372. Epub 2004 Feb 26. Mol Endocrinol. 2004. PMID: 14988428 - Two murine homologs of the Drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein.
Probst MR, Fan CM, Tessier-Lavigne M, Hankinson O. Probst MR, et al. J Biol Chem. 1997 Feb 14;272(7):4451-7. doi: 10.1074/jbc.272.7.4451. J Biol Chem. 1997. PMID: 9020169 - Cloning of two human homologs of the Drosophila single-minded gene SIM1 on chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region.
Chrast R, Scott HS, Chen H, Kudoh J, Rossier C, Minoshima S, Wang Y, Shimizu N, Antonarakis SE. Chrast R, et al. Genome Res. 1997 Jun;7(6):615-24. doi: 10.1101/gr.7.6.615. Genome Res. 1997. PMID: 9199934 Free PMC article.
Cited by
- The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation.
Wellberg E, Metz RP, Parker C, Porter WW. Wellberg E, et al. Development. 2010 Mar;137(6):945-52. doi: 10.1242/dev.041657. Epub 2010 Feb 11. Development. 2010. PMID: 20150276 Free PMC article. - Congenital and idiopathic scoliosis: clinical and genetic aspects.
Giampietro PF, Blank RD, Raggio CL, Merchant S, Jacobsen FS, Faciszewski T, Shukla SK, Greenlee AR, Reynolds C, Schowalter DB. Giampietro PF, et al. Clin Med Res. 2003 Apr;1(2):125-36. doi: 10.3121/cmr.1.2.125. Clin Med Res. 2003. PMID: 15931299 Free PMC article. Review. - Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2.
Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, Schedin P, Porter WW. Laffin B, et al. Mol Cell Biol. 2008 Mar;28(6):1936-46. doi: 10.1128/MCB.01701-07. Epub 2007 Dec 26. Mol Cell Biol. 2008. PMID: 18160708 Free PMC article. - Characterization of functionally deficient SIM2 variants found in patients with neurological phenotypes.
Button EL, Rossi JJ, McDougal DP, Bruning JB, Peet DJ, Bersten DC, Rosenfeld JA, Whitelaw ML. Button EL, et al. Biochem J. 2022 Jul 15;479(13):1441-1454. doi: 10.1042/BCJ20220209. Biochem J. 2022. PMID: 35730699 Free PMC article. - Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish.
Cheng CN, Wingert RA. Cheng CN, et al. Dev Biol. 2015 Mar 1;399(1):100-116. doi: 10.1016/j.ydbio.2014.12.020. Epub 2014 Dec 25. Dev Biol. 2015. PMID: 25542995 Free PMC article.
References
- Byragyn, A., S. Arkins, and K. W. Kelley. 1994. Riboprobe expression cassettes for measuring IGF-I, b-actin, and glyceraldehyde 3-phospate dehydrogenase transcripts. J. Immunol. Methods 168:235-244. - PubMed
- Chen, H., R. Chrast, C. Rossier, A. Gos, S. E. Antonarakis, J. Kudoh, A. Yamaki, N. Shindoh, H. Maeda, and S. Minoshima. 1995. Single-minded and Down syndrome? Nat. Genet. 10:9-10. - PubMed
- Chrast, R., H. S. Scott, H. Chen, J. Kudoh, C. Rossier, S. Minoshima, Y. Wang, N. Shimizu, and S. E. Antonarakis. 1997. Cloning of two human homologs of the Drosophila single-minded gene SIM1 on chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region. Genome Res. 7:615-624. - PMC - PubMed
- Chrast, R., H. S. Scott, R. Madani, L. Huber, D. P. Wolfer, M. Prinz, A. Aguzzi, H. P. Lipp, and S. E. Antonarakis. 2000. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9:1853-1864. - PubMed
- Crews, S., R. Franks, S. Hu, B. Matthews, and J. Nambu. 1992. Drosophila single-minded gene and the molecular genetics of CNS midline development. J. Exp. Zool. 261:234-244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases