BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets - PubMed (original) (raw)
BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets
Timothy K MacLachlan et al. Mol Cell Biol. 2002 Jun.
Abstract
The pathway leading to BRCA1-dependent tumor suppression is not yet clear but appears to involve activities in DNA repair as well as gene transcription. Moreover, it has been shown that BRCA1 can regulate p53-dependent transcription. Because BRCA1 overexpression stabilizes wild-type p53 but does not lead to apoptosis of most cell lines, we investigated the selectivity of BRCA1 for p53-dependent target gene activation. We find that BRCA1-stabilized p53 regulates transcription of DNA repair and growth arrest genes while p53 stabilized by DNA-damaging agents induces a wide array of genes, including those involved in apoptosis. This differential expression profile was reflected in the treatment outcome--apoptosis following DNA damage and growth arrest after expression of BRCA1. Depletion of BRCA1 in wild-type-p53-expressing cells abolished the induction of such repair genes as p53R2, while the expression of PIG3, an apoptosis-inducing gene, was still induced. BRCA1 also conferred diminished cell death in a p53-dependent manner in response to adriamycin compared to that conferred by controls. These results suggest that BRCA1 selectively coactivates the p53 transcription factor towards genes that direct DNA repair and cell cycle arrest but not towards those that direct apoptosis.
Figures
FIG. 1.
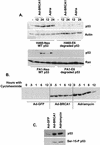
Equal stabilization of p53 by BRCA1 and adriamycin. (A) H460-Neo (wild-type-p53-expressing) and H460-E6 (degraded-p53-expressing) cells were either infected with Ad-GFP (−) or Ad-BRCA1 at an MOI of 20 or treated with 0.2 μg of adriamycin (Adria)/ml for 12 and 24 h, after which nuclear protein was harvested. The protein was immunoblotted with antibodies to p53, p21WAF1, and actin or Ran. (B) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h, treated with 10 μg of cycloheximide/ml, and then harvested for total protein at the indicated time points. Total protein was immunoblotted for p53 expression. (C) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h and then harvested for total protein. Total protein was immunoblotted for p53 and serine 15-phosphorylated (Ser-15-P) p53. WT, wild-type.
FIG. 2.
Different consequences of p53 stabilization by BRCA1 and adriamycin. (A) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 24 h. Cells were then harvested and analyzed for bromodeoxyuridine (BrdU) uptake for detection of cells in S phase. The insets in the bromodeoxyuridine FACS analyses are two-dimensional views of the propidium iodide (PI) stain from the same cells. (B) H460-Neo cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 24 h. Cells were then harvested and analyzed for the presence of active caspase 3 enzyme for the detection of cells in apoptosis. (C) PA1-Neo (wild-type-p53-expressing) and PA1-E6 (degraded-p53-expressing) cells were either infected with Ad-GFP (−) or Ad-BRCA1 at an MOI of 20 or treated with 200 ng of adriamycin/ml for 18 h, after which the cells were harvested, fixed, and stained for cellular DNA content with propidium iodide. Samples were analyzed for propidium iodide fluorescence by flow cytometry. WT, wild-type.
FIG. 3.
BRCA1 controls a subset of p53-regulated genes. H460-Neo and -E6 cells were infected with GFP (G), p53 (P), or BRCA1 (B) adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin (A)/ml for 24 h and harvested for total RNA. RNA was Northern blotted for the indicated genes. The expression pattern of the effect of BRCA1 on p53 transcriptional targets fell into three categories: genes that were affected by BRCA1 expression regardless of the status of p53 (A), genes that were affected by BRCA1 only in cells that express wild-type p53 (B), and genes that were not affected by BRCA1 expression (C). (D) SW480 cells were infected with Ad-LacZ, Ad-p53, or Ad-BRCA1 at an MOI of 20 for 24 h and then harvested for total RNA. Total RNA was Northern blotted for expression of the indicated genes. EtBr, ethidium bromide.
FIG. 3.
BRCA1 controls a subset of p53-regulated genes. H460-Neo and -E6 cells were infected with GFP (G), p53 (P), or BRCA1 (B) adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin (A)/ml for 24 h and harvested for total RNA. RNA was Northern blotted for the indicated genes. The expression pattern of the effect of BRCA1 on p53 transcriptional targets fell into three categories: genes that were affected by BRCA1 expression regardless of the status of p53 (A), genes that were affected by BRCA1 only in cells that express wild-type p53 (B), and genes that were not affected by BRCA1 expression (C). (D) SW480 cells were infected with Ad-LacZ, Ad-p53, or Ad-BRCA1 at an MOI of 20 for 24 h and then harvested for total RNA. Total RNA was Northern blotted for expression of the indicated genes. EtBr, ethidium bromide.
FIG. 3.
BRCA1 controls a subset of p53-regulated genes. H460-Neo and -E6 cells were infected with GFP (G), p53 (P), or BRCA1 (B) adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin (A)/ml for 24 h and harvested for total RNA. RNA was Northern blotted for the indicated genes. The expression pattern of the effect of BRCA1 on p53 transcriptional targets fell into three categories: genes that were affected by BRCA1 expression regardless of the status of p53 (A), genes that were affected by BRCA1 only in cells that express wild-type p53 (B), and genes that were not affected by BRCA1 expression (C). (D) SW480 cells were infected with Ad-LacZ, Ad-p53, or Ad-BRCA1 at an MOI of 20 for 24 h and then harvested for total RNA. Total RNA was Northern blotted for expression of the indicated genes. EtBr, ethidium bromide.
FIG. 3.
BRCA1 controls a subset of p53-regulated genes. H460-Neo and -E6 cells were infected with GFP (G), p53 (P), or BRCA1 (B) adenoviruses at an MOI of 20 or treated with 0.2 μg of adriamycin (A)/ml for 24 h and harvested for total RNA. RNA was Northern blotted for the indicated genes. The expression pattern of the effect of BRCA1 on p53 transcriptional targets fell into three categories: genes that were affected by BRCA1 expression regardless of the status of p53 (A), genes that were affected by BRCA1 only in cells that express wild-type p53 (B), and genes that were not affected by BRCA1 expression (C). (D) SW480 cells were infected with Ad-LacZ, Ad-p53, or Ad-BRCA1 at an MOI of 20 for 24 h and then harvested for total RNA. Total RNA was Northern blotted for expression of the indicated genes. EtBr, ethidium bromide.
FIG. 4.
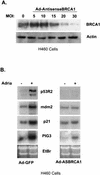
Antisense BRCA1 blocks BRCA1 protein expression and alters the induction of p53 targets. (A) H460 cells were infected with an adenovirus that expresses the first 1,500 nucleotides of BRCA1 in the antisense orientation at increasing MOI, harvested for total protein, and immunoblotted for BRCA1. (B) H460 cells were infected with Ad-AntisenseBRCA1 at an MOI of 20 or control infected and exposed to 0.2 μg of adriamycin (Adria)/ml. Total RNA was harvested 12 h later and Northern blotted for the indicated genes. EtBr, ethidium bromide.
FIG. 5.
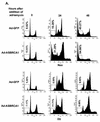
Antisense BRCA1 sensitizes cells to apoptosis stimuli. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then stained with Coomassie blue to detect attached cells after 18 h of treatment. (C) H460 and SW480 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20 or 50, respectively, and exposed to 25 J of UV/m2. Total protein was harvested at 0, 6, 12, and 18 h following irradiation and immunoblotted for caspase-3 expression, and separate wells were stained with Coomassie blue 3 days later. L, Ad-LacZ; An, Ad-Antisense BRCA1.
FIG. 5.
Antisense BRCA1 sensitizes cells to apoptosis stimuli. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20, treated with 0.2 μg of adriamycin/ml or left untreated, and then stained with Coomassie blue to detect attached cells after 18 h of treatment. (C) H460 and SW480 cells were infected with Ad-GFP or Ad-AntisenseBRCA1 at an MOI of 20 or 50, respectively, and exposed to 25 J of UV/m2. Total protein was harvested at 0, 6, 12, and 18 h following irradiation and immunoblotted for caspase-3 expression, and separate wells were stained with Coomassie blue 3 days later. L, Ad-LacZ; An, Ad-Antisense BRCA1.
FIG. 6.
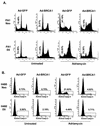
BRCA1 infection enhances cell survival in a p53-dependent manner. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis at 24 h after treatment with adriamycin. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of active caspase-3 to detect cells undergoing apoptosis. (C) PA1 and H460 cells were infected with Ad-LacZ or Ad-BRCA1 and then plated to T25 flasks at a concentration of 3 × 106 cells/flask. Cells were exposed to 10-γ gamma irradiation and then stained with Coomassie blue 1 week later. (D) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 for 12 h, treated with 200 ng of adriamycin/ml or left untreated, and then harvested for total RNA at 2, 4, and 6 h after adriamycin treatment. RNA was blotted for the expression of the indicated genes. EtBr, ethidium bromide.
FIG. 6.
BRCA1 infection enhances cell survival in a p53-dependent manner. (A) PA1-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of cells containing less than 2N genomic DNA content by FACS analysis at 24 h after treatment with adriamycin. (B) H460-Neo and -E6 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20, treated with 200 ng of adriamycin/ml or left untreated, and then measured for the presence of active caspase-3 to detect cells undergoing apoptosis. (C) PA1 and H460 cells were infected with Ad-LacZ or Ad-BRCA1 and then plated to T25 flasks at a concentration of 3 × 106 cells/flask. Cells were exposed to 10-γ gamma irradiation and then stained with Coomassie blue 1 week later. (D) H460 cells were infected with Ad-GFP or Ad-BRCA1 at an MOI of 20 for 12 h, treated with 200 ng of adriamycin/ml or left untreated, and then harvested for total RNA at 2, 4, and 6 h after adriamycin treatment. RNA was blotted for the expression of the indicated genes. EtBr, ethidium bromide.
Similar articles
- Impairment of the DNA repair and growth arrest pathways by p53R2 silencing enhances DNA damage-induced apoptosis in a p53-dependent manner in prostate cancer cells.
Devlin HL, Mack PC, Burich RA, Gumerlock PH, Kung HJ, Mudryj M, deVere White RW. Devlin HL, et al. Mol Cancer Res. 2008 May;6(5):808-18. doi: 10.1158/1541-7786.MCR-07-2027. Mol Cancer Res. 2008. PMID: 18505925 - Expression and mutation analyses of P53R2, a newly identified p53 target for DNA repair in human gastric carcinoma.
Byun DS, Chae KS, Ryu BK, Lee MG, Chi SG. Byun DS, et al. Int J Cancer. 2002 Apr 10;98(5):718-23. doi: 10.1002/ijc.10253. Int J Cancer. 2002. PMID: 11920641 - Repression of BRCA1 through a feedback loop involving p53.
MacLachlan TK, Dash BC, Dicker DT, El-Deiry WS. MacLachlan TK, et al. J Biol Chem. 2000 Oct 13;275(41):31869-75. doi: 10.1074/jbc.M003338200. J Biol Chem. 2000. PMID: 10884389 - Transactivation of repair genes by BRCA1.
El-Deiry WS. El-Deiry WS. Cancer Biol Ther. 2002 Sep-Oct;1(5):490-1. doi: 10.4161/cbt.1.5.162. Cancer Biol Ther. 2002. PMID: 12496474 Review. - BRCA1 regulation of transcription.
Rosen EM, Fan S, Ma Y. Rosen EM, et al. Cancer Lett. 2006 May 18;236(2):175-85. doi: 10.1016/j.canlet.2005.04.037. Epub 2005 Jun 21. Cancer Lett. 2006. PMID: 15975711 Review.
Cited by
- Differential effects of HTLV-1 Tax oncoprotein on the different estrogen-induced-ER α-mediated transcriptional activities.
Abou-Kandil A, Eisa N, Jabareen A, Huleihel M. Abou-Kandil A, et al. Cell Cycle. 2016 Oct;15(19):2626-2635. doi: 10.1080/15384101.2016.1208871. Epub 2016 Jul 15. Cell Cycle. 2016. PMID: 27420286 Free PMC article. - BRCA1-Dependent Transcriptional Regulation: Implication in Tissue-Specific Tumor Suppression.
Zhang X, Li R. Zhang X, et al. Cancers (Basel). 2018 Dec 14;10(12):513. doi: 10.3390/cancers10120513. Cancers (Basel). 2018. PMID: 30558184 Free PMC article. Review. - Microarray-based transcriptional profiling of Eimeria bovis-infected bovine endothelial host cells.
Taubert A, Wimmers K, Ponsuksili S, Jimenez CA, Zahner H, Hermosilla C. Taubert A, et al. Vet Res. 2010 Sep-Oct;41(5):70. doi: 10.1051/vetres/2010041. Epub 2010 Jul 12. Vet Res. 2010. PMID: 20615380 Free PMC article. - BRCA1 inhibits membrane estrogen and growth factor receptor signaling to cell proliferation in breast cancer.
Razandi M, Pedram A, Rosen EM, Levin ER. Razandi M, et al. Mol Cell Biol. 2004 Jul;24(13):5900-13. doi: 10.1128/MCB.24.13.5900-5913.2004. Mol Cell Biol. 2004. PMID: 15199145 Free PMC article. - Mathematical model of dynamic protein interactions regulating p53 protein stability for tumor suppression.
Wang H, Peng G. Wang H, et al. Comput Math Methods Med. 2013;2013:358980. doi: 10.1155/2013/358980. Epub 2013 Dec 28. Comput Math Methods Med. 2013. PMID: 24454532 Free PMC article.
References
- Abbott, D. W., M. E. Thompson, C. Robinson-Benion, G. Tomlinson, R. A. Jensen, and J. T. Holt. 1999. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 274:18808-18812. - PubMed
- Bates, S., S. Rowan, and K. H. Vousden. 1996. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13:1103-1109. - PubMed
- Chen, J., D. P. Silver, D. Walpita, S. B. Cantor, A. F. Gazdar, G. Tomlinson, F. J. Couch, B. L. Weber, T. Ashley, D. M. Livingston, and R. Scully. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317-328. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous