Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression - PubMed (original) (raw)
Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression
Gerda Lagger et al. EMBO J. 2002.
Abstract
Histone deacetylases (HDACs) modulate chromatin structure and transcription, but little is known about their function in mammalian development. HDAC1 was implicated previously in the repression of genes required for cell proliferation and differentiation. Here we show that targeted disruption of both HDAC1 alleles results in embryonic lethality before E10.5 due to severe proliferation defects and retardation in development. HDAC1-deficient embryonic stem cells show reduced proliferation rates, which correlate with decreased cyclin-associated kinase activities and elevated levels of the cyclin-dependent kinase inhibitors p21(WAF1/CIP1) and p27(KIP1). Similarly, expression of p21 and p27 is up-regulated in HDAC1-null embryos. In addition, loss of HDAC1 leads to significantly reduced overall deacetylase activity, hyperacetylation of a subset of histones H3 and H4 and concomitant changes in other histone modifications. The expression of HDAC2 and HDAC3 is induced in HDAC1-deficient cells, but cannot compensate for loss of the enzyme, suggesting a unique function for HDAC1. Our study provides the first evidence that a histone deacetylase is essential for unrestricted cell proliferation by repressing the expression of selective cell cycle inhibitors.
Figures
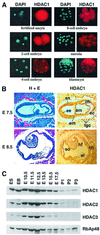
Fig. 1. HDAC1 expression during mouse embryonic development. (A) HDAC1 expression in pre-implantation embryos from the fertilized oocyte to the blastocyst stage was monitored in indirect immunofluorescence experiments. HDAC1 was visualized using a polyclonal HDAC1 antiserum and a secondary Texas red-conjugated antibody. Genomic DNA was stained with DAPI. (B) Immunohistochemical analysis of HDAC1 expression in paraffin-embedded sections of mouse embryos. Adjacent sections were stained with hematoxylin/eosin (H+E) or immunostained for HDAC1 at E7.5 (upper panels) and E8.5 (lower panels). High HDAC1 expression is found in all embryonic and extra-embryonic tissues, in particular in trophoblast giant cells. am, amnion; ch, chorion; ec, ectoplacental cone; ee, embryonic ectoderm; em, embryonic mesoderm; en, endoderm; tgc, trophoblast giant cells; al, allantois; hf, headfold; so, somites. (C) Expression patterns of HDAC1, HDAC2, HDAC3 and RbAp48 during mouse embryogenesis. Western blot analysis of whole-cell extracts from ES cells, embryoid bodies (EB) and 10.5- to 17.5-day-old embryos (E10.5–E17.5). P1-3 indicates 1–3 days after birth. Exposure times for ECL detection were adjusted approximately to the individual sensitivities of the HDAC antibodies.
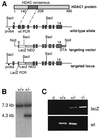
Fig. 2. Disruption of the murine HDAC1 locus. (A) Structure of the mouse Hdac1 gene locus. In the targeting vector, part of exon 5 and exons 6 and 7 encoding the deacetylase consensus motif were replaced by the lacZ/neo cassette for G418 selection of positive clones. The diphtheria toxin gene (DTA) was inserted 3′ of the long arm to select against random integration of the targeting construct. (B) Southern blot analysis of tail DNA isolated from offspring of heterozygous intercrosses. Genomic DNA was digested with _Sac_I and hybridized with an intron-specific probe, which recognizes a 7.3 kb fragment in the wild-type allele and a 4.3 kb S_ac_I fragment in the targeted locus. (C) Yolk sac PCR analysis of E9.5 embryos.
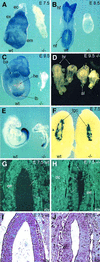
Fig. 3. Phenotypic analysis of HDAC1 mutant embryos. (A–D) Whole-mount in situ hybridization of wild-type and HDAC1 mutant embryos with an HDAC1 riboprobe. (A) HDAC1 is highly expressed in the ectoplacental cone (ec), the extra-embryonic (ex) and embryonic (em) ectoderm at E7.5. (B) Elevated HDAC1 levels are detected in the head fold (hf) and the neural fold (nf) at E8.5. (C) HDAC1 is highly expressed throughout the E9.5 embryo except in the developing heart (he), with pronounced expression in the bronchial arches (ba) and the limb bud (lb). (D) Spectrum of phenotypes observed for HDAC1 mutant embryos at E9.5 with misformed allantois (al) and defects in head formation; hr, head region. (E) Brachyury expression at E9.5 is comparable in HDAC1 wild-type and HDAC1 mutant embryos. (F) E7.5 wild-type and mutant embryos within the maternal decidua are shown. Placental lactogen-1 expression in trophoblast giant cells (tgc) is similar in HDAC1 wild-type and HDAC1 null embryos. (G and H) TUNEL assay on paraffin-embedded sections of E7.5 wild-type (G) and mutant (H) embryos shows similar numbers of apoptotic cells in the embryonic ectoderm (ee) of wild-type and mutant embryos. Arrows indicate apoptotic cells. (I and J) Hematoxylin/eosin staining of sections adjacent to the sections shown in (G) and (H).
Fig. 4. Reduced proliferation of HDAC1-null embryos. Immunohisto chemical analysis of HDAC1 and Ki67 antigen expression in mouse embryos at E7.5. Adjacent paraffin-embedded sagittal sections of mouse embryos were stained with hematoxylin/eosin (A and B) or immunostained for HDAC1 (C and D) or for the proliferation marker Ki67 nuclear antigen (E and F). The boxed areas show higher magnifications of the Ki67-stained embryo sections.
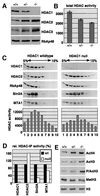
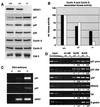
Fig. 5. HDAC1-deficient ES cells display decreased histone deacetylase activity and changes in histone modifications. (A) Western blot analysis of homologous HDAC proteins of total protein extracts prepared from wild-type, heterozygous and null ES cell lines. The blot was incubated sequentially with antibodies directed against HDAC1, HDAC2, HDAC3 and RbAp48, respectively. (B) Equal amounts of extracts described in (A) were analyzed for deacetylase activity with tritium acetate-labeled histones as substrates. Counted radioactivity corresponds to the amount of released acetyl moieties per hour and 10 µg protein and reflects the relative HDAC activity. Results are shown as mean values of three independent experiments. (C) Co-sedimentation analysis of HDAC1–HDAC2-containing complexes in wild-type and HDAC1-null cells. One aliquot of each fraction was analyzed on western blots for the presence of components of HDAC1–HDAC2 complexes. A second aliquot of each fraction was tested for total HDAC activity. (D) Comparison of deacetylase activities associated with components of the HDAC1–HDAC2 complexes. HDAC1, HDAC2, Sin3A and MTA1 were immunoprecipitated from whole-cell extracts prepared from wild-type or HDAC1-null ES cells and analyzed for associated HDAC activity as described in (B). The data shown are representative of three independent experiments. (E) Changes in core histone modifications in HDAC1 heterozygous and homozygous ES cells. Lack of HDAC1 resulted in H3 and H4 hyperacetylation, increased S10/K14 phosphoacetylation and reduced K9 methylation of histone H3. Histones were extracted and analyzed on western blots with antibodies recognizing acetyl-histone H4 (AcH4), acetyl-histone H3 (AcH3), histone H3-phosphoS10-acetylK14 (P/AcH3) and histone H3-methylK9 (MetH3). Equal loading was controlled by probing with an H3 antibody (H3).
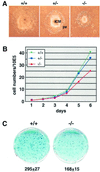
Fig. 6. Loss of HDAC1 in ES cells leads to impaired proliferation. (A) Three ES cell lines of different genotype were subjected to differentiation. The HDAC1-null embryoid bodies show a reduced inner cell mass (ICM), but normal differentiation. pe, parietal endoderm. (B) Growth curves of HDAC1 wild-type, heterozygous and null ES cell lines. Equal numbers of cells (5 × 105) were seeded in triplicate and aliquots were counted daily during a time period of 6 days. (C) Colony formation assay with wild-type and HDAC1 mutant ES cells. A total of 1.5 × 103 cells were seeded on SNL feeder layers and cultivated for 8 days. Cells were fixed and stained with methylene blue and colonies were counted. The average clone numbers of four plates are indicated with the respective standard deviations.
Fig. 7. The CDK inhibitors p21 and p27 are up-regulated in HDAC1-deficient ES cells. (A) Western blot analysis of whole-cell extracts prepared from wild-type, heterozygous and null ES cell lines. Western blots were probed sequentially with the indicated antibodies. (B) HDAC1-deficient ES cells show reduced cyclin A- and cyclin E-associated kinase activity. Cyclin A or cyclin E complexes were immunoprecipitated from ES cell extracts and incubated with histone H1 in the presence of [γ-32P]ATP. Phospholabeled histone H1 was quantified and is shown for cyclin A (black bars) and cyclin E (white bars) relative to the kinase activity of wild-type cells arbitrarily set as 100. The results are representative of three independent experiments. (C) p21 and p27 expression is increased in E9.5 mutant embryos. HPRT was used as a control. The levels of p21, p27 and HPRT mRNA were determined by semiquantitative RT–PCR analysis. (D) Hyperacetylation of specific target promoters in HDAC1-null cells. Chromatin isolated from wild-type or HDAC1-deficient cells was precipitated in the absence of specific antibodies (no AB) or with antibodies specific for acetylated histone H3 (AcH3) or acetylated histone H4 (AcH4). Total input DNA (1×, 0.25× and 0.0625×) and DNA from the antibody-bound fractions were analyzed by quantitative PCR. Amounts of amplified DNA were quantified and are indicated relative to the signals from wild-type cells (set to 1).
Similar articles
- Che-1 arrests human colon carcinoma cell proliferation by displacing HDAC1 from the p21WAF1/CIP1 promoter.
Di Padova M, Bruno T, De Nicola F, Iezzi S, D'Angelo C, Gallo R, Nicosia D, Corbi N, Biroccio A, Floridi A, Passananti C, Fanciulli M. Di Padova M, et al. J Biol Chem. 2003 Sep 19;278(38):36496-504. doi: 10.1074/jbc.M306694200. Epub 2003 Jul 7. J Biol Chem. 2003. PMID: 12847090 - The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene.
Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G, Rotheneder H, Wintersberger E, Seiser C. Lagger G, et al. Mol Cell Biol. 2003 Apr;23(8):2669-79. doi: 10.1128/MCB.23.8.2669-2679.2003. Mol Cell Biol. 2003. PMID: 12665570 Free PMC article. - Cyclin-dependent kinase inhibitors, p21(waf1/cip1) and p27(kip1), are expressed site- and hair cycle-dependently in rat hair follicles.
Mitsui S, Ohuchi A, Adachi-Yamada T, Hotta M, Tsuboi R, Ogawa H. Mitsui S, et al. J Dermatol Sci. 2001 Feb;25(2):164-9. doi: 10.1016/s0923-1811(00)00132-8. J Dermatol Sci. 2001. PMID: 11164713 - Histone deacetylase inhibitors: signalling towards p21cip1/waf1.
Ocker M, Schneider-Stock R. Ocker M, et al. Int J Biochem Cell Biol. 2007;39(7-8):1367-74. doi: 10.1016/j.biocel.2007.03.001. Epub 2007 Mar 7. Int J Biochem Cell Biol. 2007. PMID: 17412634 Review.
Cited by
- Selective class I histone deacetylase inhibition suppresses hypoxia-induced cardiopulmonary remodeling through an antiproliferative mechanism.
Cavasin MA, Demos-Davies K, Horn TR, Walker LA, Lemon DD, Birdsey N, Weiser-Evans MC, Harral J, Irwin DC, Anwar A, Yeager ME, Li M, Watson PA, Nemenoff RA, Buttrick PM, Stenmark KR, McKinsey TA. Cavasin MA, et al. Circ Res. 2012 Mar 2;110(5):739-48. doi: 10.1161/CIRCRESAHA.111.258426. Epub 2012 Jan 26. Circ Res. 2012. PMID: 22282194 Free PMC article. - Disrupted ectodermal organ morphogenesis in mice with a conditional histone deacetylase 1, 2 deletion in the epidermis.
Hughes MW, Jiang TX, Lin SJ, Leung Y, Kobielak K, Widelitz RB, Chuong CM. Hughes MW, et al. J Invest Dermatol. 2014 Jan;134(1):24-32. doi: 10.1038/jid.2013.283. Epub 2013 Jun 21. J Invest Dermatol. 2014. PMID: 23792463 Free PMC article. - Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation.
Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han MH, Bassel-Duby R, Olson EN, Nestler EJ. Kennedy PJ, et al. Nat Neurosci. 2013 Apr;16(4):434-40. doi: 10.1038/nn.3354. Epub 2013 Mar 10. Nat Neurosci. 2013. PMID: 23475113 Free PMC article. - Lentivirus-mediated Knockdown of HDAC1 Uncovers Its Role in Esophageal Cancer Metastasis and Chemosensitivity.
Song M, He G, Wang Y, Pang X, Zhang B. Song M, et al. J Cancer. 2016 Jul 26;7(12):1694-1700. doi: 10.7150/jca.15086. eCollection 2016. J Cancer. 2016. PMID: 27698906 Free PMC article. - Modulation of cell cycle regulators by HDACs.
Telles E, Seto E. Telles E, et al. Front Biosci (Schol Ed). 2012 Jan 1;4(3):831-9. doi: 10.2741/s303. Front Biosci (Schol Ed). 2012. PMID: 22202094 Free PMC article. Review.
References
- Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet., 16, 351–356. - PubMed
- Bader A., Al Dubai,H. and Weitzer,G. (2000) Leukemia inhibitory factor modulates cardiogenesis in embryoid bodies in opposite fashions. Circ. Res., 86, 787–794. - PubMed
- Brown J.L., Mucci,D., Whiteley,M., Dirksen,M.L. and Kassis,J.A. (1998) The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell, 1, 1057–1064. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous