Systematic sequencing of cDNA clones using the transposon Tn5 - PubMed (original) (raw)
Systematic sequencing of cDNA clones using the transposon Tn5
Yuriy Shevchenko et al. Nucleic Acids Res. 2002.
Abstract
In parallel with the production of genomic sequence data, attention is being focused on the generation of comprehensive cDNA-sequence resources. Such efforts are increasingly emphasizing the production of high-accuracy sequence corresponding to the entire insert of cDNA clones, especially those presumed to reflect the full-length mRNA. The complete sequencing of cDNA clones on a large scale presents unique challenges because of the generally small, yet heterogeneous, sizes of the cloned inserts. We have developed a strategy for high-throughput sequencing of cDNA clones using the transposon Tn5. This approach has been tailored for implementation within an existing large-scale 'shotgun-style' sequencing program, although it could be readily adapted for use in virtually any sequencing environment. In addition, we have developed a modified version of our strategy that can be applied to cDNA clones with large cloning vectors, thereby overcoming a potential limitation of transposon-based approaches. Here we describe the details of our cDNA-sequencing pipeline, including a summary of the experience in sequencing more than 4200 cDNA clones to produce more than 8 million base pairs of high-accuracy cDNA sequence. These data provide both convincing evidence that the insertion of Tn5 into cDNA clones is sufficiently random for its effective use in large-scale cDNA sequencing as well as interesting insight about the sequence context preferred for insertion by Tn5.
Figures
Figure 1
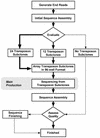
Pipeline for transposon-based sequencing of cDNA clones. The general pipeline for the systematic sequencing of cDNA clones using the transposon Tn5 is depicted, with additional details provided in the text. Note that ‘transposon subclones’ correspond to subclones derived from the starting cDNA clone by the insertion of a transposon. For some steps, the thickness of the arrows is intended to reflect the relative number of cDNA clones traversing that portion of the pipeline (see Table 1). The gray box designates the portion of the pipeline that can be readily performed as part of the ‘main production’ component of a DNA-sequencing facility.
Figure 2
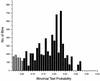
Assessing randomness of transposon Tn5 insertions. The binomial test was used to assess the distribution of transposon-insertion events. The insertions of Tn5 into 1955 cDNA clones were analyzed and assigned to bins (see Materials and Methods for details). The resulting _P_-values reflect the likelihood that the observed insertion events were not random. Plotted are the numbers of bins grouped into _P_-value ranges of 0.01. _P_-values >0.05 correspond to bins for which the observed insertion events are likely to be random. _P_-values ≤0.05 (indicated by gray bars) correspond to bins for which the observed insertion events cannot be confidently described as random occurrences.
Figure 3
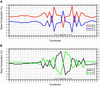
Base composition at the Tn5-insertion site and immediately flanking it. The frequency of each base flanking 24 493 Tn5-insertion events was cataloged (see Table 2). From those data, the relative compositions of GC and AT (A) and pyrimidine (Py) and purine (Pu) nucleotides (B) were determined and then plotted relative to the 9-bp target site, where position 1 is the 5′ end of the site.
Figure 4
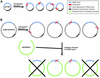
Modified strategy for transposon-based sequencing of cDNA clones involving Gateway cloning technology. The transposon-based approach for sequencing cDNA clones described here can be implemented in the most straightforward fashion with clones containing relatively small vectors, such as pOTB7 (A). In these cases, most of the resulting transposon-containing subclones harbor a transposon within the cDNA insert. While insertions within the vector backbone occur, those inserting within the essential components of the vector (e.g. antibiotic resistance gene, origin of replication) yield non-viable subclones; thus, only a small minority of the recovered subclones harbor a transposon in the vector. For cDNA clones with larger vectors, such as pCMV-SPORT6.0 (B), a much larger proportion of transposon-insertion events occur within the vector backbone, with only a small fraction occurring within the essential components of the vector. Undesirable ‘background’ subclones (i.e. those with an inserted transposon in the vector) can be eliminated by using the Gateway-transfer system (27,28) to shuttle the cDNA inserts into a suitable recipient vector (e.g. pDONR223). By then selecting for the recipient vector backbone and the presence of a transposon, virtually all of the resulting subclones should harbor a transposon within the transferred cDNA insert. Subclones containing a cDNA insert devoid of a transposon would be non-viable (indicated by crosses). Note that the vectors and cDNA inserts are not drawn to scale. At both ends of each inserted transposon are annealing sites for sequencing primers (arrows).
Similar articles
- An efficient strategy for large-scale high-throughput transposon-mediated sequencing of cDNA clones.
Butterfield YS, Marra MA, Asano JK, Chan SY, Guin R, Krzywinski MI, Lee SS, MacDonald KW, Mathewson CA, Olson TE, Pandoh PK, Prabhu AL, Schnerch A, Skalska U, Smailus DE, Stott JM, Tsai MI, Yang GS, Zuyderduyn SD, Schein JE, Jones SJ. Butterfield YS, et al. Nucleic Acids Res. 2002 Jun 1;30(11):2460-8. doi: 10.1093/nar/30.11.2460. Nucleic Acids Res. 2002. PMID: 12034834 Free PMC article. - A rapid and cost-effective method for sequencing pooled cDNA clones by using a combination of transposon insertion and Gateway technology.
Morozumi T, Toki D, Eguchi-Ogawa T, Uenishi H. Morozumi T, et al. Biotechniques. 2011 Sep;51(3):195-7. doi: 10.2144/000113737. Biotechniques. 2011. PMID: 21906043 - Tn5 as a molecular genetics tool: In vitro transposition and the coupling of in vitro technologies with in vivo transposition.
Reznikoff WS, Goryshin IY, Jendrisak JJ. Reznikoff WS, et al. Methods Mol Biol. 2004;260:83-96. doi: 10.1385/1-59259-755-6:083. Methods Mol Biol. 2004. PMID: 15020804 - Large-scale EST sequencing in rice.
Yamamoto K, Sasaki T. Yamamoto K, et al. Plant Mol Biol. 1997 Sep;35(1-2):135-44. Plant Mol Biol. 1997. PMID: 9291967 Review. - Optical mapping and its potential for large-scale sequencing projects.
Aston C, Mishra B, Schwartz DC. Aston C, et al. Trends Biotechnol. 1999 Jul;17(7):297-302. doi: 10.1016/s0167-7799(99)01326-8. Trends Biotechnol. 1999. PMID: 10370237 Review.
Cited by
- Tagmentation-based analysis reveals the clonal behavior of CAR-T cells in association with lentivector integration sites.
Kim J, Park M, Baek G, Kim JI, Kwon E, Kang BC, Kim JI, Kang HJ. Kim J, et al. Mol Ther Oncolytics. 2023 May 16;30:1-13. doi: 10.1016/j.omto.2023.05.004. eCollection 2023 Sep 21. Mol Ther Oncolytics. 2023. PMID: 37360944 Free PMC article. - Enabling low-cost and robust essentiality studies with high-throughput transposon mutagenesis (HTTM).
Champie A, De Grandmaison A, Jeanneau S, Grenier F, Jacques PÉ, Rodrigue S. Champie A, et al. PLoS One. 2023 Apr 11;18(4):e0283990. doi: 10.1371/journal.pone.0283990. eCollection 2023. PLoS One. 2023. PMID: 37040373 Free PMC article. - Elucidation of Essential Genes and Mutant Fitness during Adaptation toward Nitrogen Fixation Conditions in the Endophyte Azoarcus olearius BH72 Revealed by Tn-Seq.
Harten T, Nimzyk R, Gawlick VEA, Reinhold-Hurek B. Harten T, et al. Microbiol Spectr. 2022 Dec 21;10(6):e0216222. doi: 10.1128/spectrum.02162-22. Epub 2022 Nov 23. Microbiol Spectr. 2022. PMID: 36416558 Free PMC article. - Comprehensive understanding of Tn5 insertion preference improves transcription regulatory element identification.
Zhang H, Lu T, Liu S, Yang J, Sun G, Cheng T, Xu J, Chen F, Yen K. Zhang H, et al. NAR Genom Bioinform. 2021 Oct 27;3(4):lqab094. doi: 10.1093/nargab/lqab094. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34729473 Free PMC article. - Fast and inexpensive whole-genome sequencing library preparation from intact yeast cells.
Vonesch SC, Li S, Szu Tu C, Hennig BP, Dobrev N, Steinmetz LM. Vonesch SC, et al. G3 (Bethesda). 2021 Jan 18;11(1):jkaa009. doi: 10.1093/g3journal/jkaa009. G3 (Bethesda). 2021. PMID: 33561223 Free PMC article.
References
- Green E.D. (2001) Strategies for the systematic sequencing of complex genomes. Nature Rev. Genet., 2, 573–583. - PubMed
- Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. - PubMed
- Hillier L., Lennon,G., Becker,M., Bonaldo,M.F., Chiapelli,B., Chissoe,S., Dietrich,N., DuBuque,T., Favello,A., Gish,W. et al. (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res., 6, 807–828. - PubMed
- Marra M., Hillier,L., Kucaba,T., Allen,M., Barstead,R., Beck,C., Blistain,A., Bonaldo,M., Bowers,Y., Bowles,L. et al. (1999) An encyclopedia of mouse genes. Nature Genet., 21, 191–194. - PubMed
- Marra M.A., Hillier,L. and Waterston,R.H. (1998) Expressed sequence tags—ESTablishing bridges between genomes. Trends Genet., 14, 4–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources