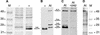Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant - PubMed (original) (raw)
Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant
Paul B Talbert et al. Plant Cell. 2002 May.
Abstract
Centromeric H3-like histones, which replace histone H3 in the centromeric chromatin of animals and fungi, have not been reported in plants. We identified a histone H3 variant from Arabidopsis thaliana that encodes a centromere-identifying protein designated HTR12. By immunological detection, HTR12 localized at centromeres in both mitotic and meiotic cells. HTR12 signal revealed tissue- and stage-specific differences in centromere morphology, including a distended bead-like structure in interphase root tip cells. The anti-HTR12 antibody also detected spherical organelles in meiotic cells. Although the antibody does not label centromeres in the closely related species Arabidopsis arenosa, HTR12 signal was found on all centromeres in allopolyploids of these two species. Comparison of the HTR12 genes of A. thaliana and A. arenosa revealed striking adaptive evolution in the N-terminal tail of the protein, similar to the pattern seen in its counterpart in Drosophila. This finding suggests that the same evolutionary forces shape centromeric chromatin in both animals and plants.
Figures
Figure 1.
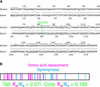
HTR12 Genes from A. thaliana and A. arenosa. (A) Predicted protein sequences of HTR12 from A. thaliana (Athal) and A. arenosa (Aare). Amino acid identities (:) and conservative substitutions (.) are indicated. Underlines and overlines indicate amino acid identity to A. thaliana histone H3. (B) Positions of nucleotide substitutions in the A. arenosa HTR12 gene that result in nonsynonymous (red) and synonymous (blue) codon changes relative to the A. thaliana gene.
Figure 2.
Protein Gel Blot Analysis Using the Anti-HTR12 Antibody. Sources of protein extracts are uninduced E. coli cells carrying the HTR12 cDNA under the control of an isopropylthio-β-galactoside–inducible T7 promoter (−), induced cells of same culture (+), and A. thaliana (At) immature inflorescences. M, molecular mass markers. Protein sizes are in kD. (A) Coomassie blue–stained SDS-PAGE gel showing the induction of HTR12 protein expression in E. coli. (B) Anti-HTR12 antibody detection on a protein gel blot comparing bacterially expressed HTR12 protein with proteins detected in A. thaliana. (C) Anti-HTR12 antibody binding to identical lanes of a protein gel blot in the absence (left) or presence (right; +pep) of the octadecapeptide antigen at a peptide:antibody molar ratio of 1:2. (D) Coomassie blue–stained SDS-PAGE gel of A. thaliana protein extract.
Figure 3.
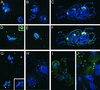
HTR12 Protein in Mitotic Tapetum Cells. Green indicates anti-HTR12; blue indicates DAPI. (A) Mitotic interphase in a premeiotic anther. (B) to (D) Mitotic cells in a meiotic anther: metaphase (center) and interphase (bottom right) (B); anaphase (center) and interphase (left) (C); and interphase binucleate cells (D). Note that in (D) the nuclei in the lower two cells are diploid, whereas those in the upper cell are tetraploid.
Figure 4.
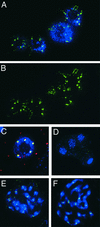
Anti-HTR12 Staining in PMCs and Pollen Cells. Green indicates anti-HTR12; blue indicates DAPI. (A) Leptotene PMC at top left, somatic cell at bottom right. (B) Zygotene and pachytene PMCs with somatic cells at bottom left. (C) and (F) Pachytene chromosomes at two different pixel intensities. In (F), note the presence of signal on the euchromatic arms but its absence from the nucleolus organizers (DAPI-bright regions on the ends of the chromosomes seen at left) and the pericentric heterochromatin. (D) and (E) Metaphase 1. Inset in (D) shows an enlargement of a niu body. (G) Anaphase 1. There is no detectable DAPI signal under niu bodies (inset). (H) Two cells of a meiotic tetrad. Note the location of niu bodies outside the nuclei. (I) Binucleate tapetum cell with fluorescent vesicles and an intact pollen grain, which fluoresces in the green channel. Anti-HTR12 antibody does not penetrate the pollen wall. (J) Crushed pollen grains and somatic cells. Strong anti-HTR12 staining in the pollen cells contrasts with smaller signals in some somatic cells (bottom left) and weak or undetectable signals in others (bottom right).
Figure 5.
Anti-HTR12 Staining in Interphase Root Tips and Allotetraploids, and Colocalization with 180-bp Repeats. (A) and (B) Anti-HTR12 staining in interphase root tip cells. DAPI staining (A) is omitted in (B) for greater visibility of the centromeric structure. (C) A probe for the 180-bp repeat (red) and anti-HTR12 staining (green) coincide or overlap (yellow) in a pollen cell nucleus. (D) Anti-HTR12 antibody detects only nonspecific background in a meiotic tetrad of A. arenosa. (E) Anti-HTR12 staining in mitotic prophase cells of a synthetic allotetraploid of A. thaliana and A. arenosa. (F) Anti-HTR12 staining in mitotic prophase cells of A. suecica.
Similar articles
- Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences.
Kawabe A, Nasuda S, Charlesworth D. Kawabe A, et al. Genetics. 2006 Dec;174(4):2021-32. doi: 10.1534/genetics.106.063628. Epub 2006 Oct 8. Genetics. 2006. PMID: 17028323 Free PMC article. - Adaptive evolution of the histone fold domain in centromeric histones.
Cooper JL, Henikoff S. Cooper JL, et al. Mol Biol Evol. 2004 Sep;21(9):1712-8. doi: 10.1093/molbev/msh179. Epub 2004 Jun 2. Mol Biol Evol. 2004. PMID: 15175412 - Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana.
Shibata F, Murata M. Shibata F, et al. J Cell Sci. 2004 Jun 15;117(Pt 14):2963-70. doi: 10.1242/jcs.01144. Epub 2004 May 25. J Cell Sci. 2004. PMID: 15161939 - Centromeres Drive a Hard Bargain.
Rosin LF, Mellone BG. Rosin LF, et al. Trends Genet. 2017 Feb;33(2):101-117. doi: 10.1016/j.tig.2016.12.001. Epub 2017 Jan 7. Trends Genet. 2017. PMID: 28069312 Free PMC article. Review. - Plant centromeres: genetics, epigenetics and evolution.
Oliveira LC, Torres GA. Oliveira LC, et al. Mol Biol Rep. 2018 Oct;45(5):1491-1497. doi: 10.1007/s11033-018-4284-7. Epub 2018 Aug 16. Mol Biol Rep. 2018. PMID: 30117088 Review.
Cited by
- Adaptive Evolution of CENP-A in Percid Fishes.
Abbey HN, Kral LG. Abbey HN, et al. Genes (Basel). 2015 Jul 17;6(3):662-71. doi: 10.3390/genes6030662. Genes (Basel). 2015. PMID: 26193324 Free PMC article. - Sequential de novo centromere formation and inactivation on a chromosomal fragment in maize.
Liu Y, Su H, Pang J, Gao Z, Wang XJ, Birchler JA, Han F. Liu Y, et al. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):E1263-71. doi: 10.1073/pnas.1418248112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733907 Free PMC article. - Chromosome dynamics visualized with an anti-centromeric histone H3 antibody in Allium.
Nagaki K, Yamamoto M, Yamaji N, Mukai Y, Murata M. Nagaki K, et al. PLoS One. 2012;7(12):e51315. doi: 10.1371/journal.pone.0051315. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236469 Free PMC article. - Structural variation and DNA methylation shape the centromere-proximal meiotic crossover landscape in Arabidopsis.
Fernandes JB, Naish M, Lian Q, Burns R, Tock AJ, Rabanal FA, Wlodzimierz P, Habring A, Nicholas RE, Weigel D, Mercier R, Henderson IR. Fernandes JB, et al. Genome Biol. 2024 Jan 22;25(1):30. doi: 10.1186/s13059-024-03163-4. Genome Biol. 2024. PMID: 38254210 Free PMC article. - Molecular and functional dissection of the maize B chromosome centromere.
Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang J. Jin W, et al. Plant Cell. 2005 May;17(5):1412-23. doi: 10.1105/tpc.104.030643. Epub 2005 Apr 1. Plant Cell. 2005. PMID: 15805482 Free PMC article.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
- Aragón-Alcaide, L., Reader, S., Miller, T., and Moore, G. (1997). Centromeric behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma 106, 327–333. - PubMed
- Bedinger, P.A., Hardeman, K.J., and Loukides, C.A. (1994). Travelling in style: The cell biology of pollen. Trends Cell Biol. 4, 132–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases