Splitting p63 - PubMed (original) (raw)
Review
Splitting p63
Hans van Bokhoven et al. Am J Hum Genet. 2002 Jul.
Erratum in
- Am J Hum Genet. 2003 Mar;72(3):779
Abstract
Causative TP63 mutations have been identified in five distinct human developmental disorders that are characterized by various degrees of limb abnormalities, ectodermal dysplasia, and facial clefts. The distribution of mutations over the various p63 protein domains and the structural and functional implications of these mutations establish a clear genotype-phenotype correlation.
Figures
Figure 1
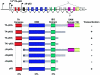
Structure of both TP63 and major protein isotypes. TP63 uses several transcription initiation sites (arrows) and extensive alternative splicing, to generate a bewildering number of different mRNAs. For clarity, several alternative-splicing routes at the 5′ end of the gene have not been indicated. Several protein domains can be distinguished; of these, the TA domains, the DBD, and the ISO domain are highly homologous to the corresponding domains in p53. The SAM domain and the TID are not contained in the p53 protein. The capacity to transactivate gene expression at a p53-responsive target is given for each of the indicated isotypes.
Figure 2
Distribution of missense mutations in the DBDs of p53 and p63. Amino acid–sequence comparison of the DBDs of p53, p63, and p73 reveals a high conservation, especially in the core domains (boxed). Within these core domains, amino acids that directly interact with the DNA and amino acids that form the Zn-binding pocket of the protein are highlighted. The black bars on top represent the incidence of somatic p53 mutations in tumors at the corresponding amino acids; the colored bars beneath the sequences represent the incidence of p63 mutations in the indicated human disorders. The p53-mutation spectrum is based on the IARC TP53 Mutation Database.
Figure 3
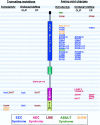
Distribution of mutations in p63, revealing a genotype-phenotype correlation. The approximate positions of truncating mutations (left) and amino acid changes (right) are indicated, together with the associated phenotype, with respect to the occurrence of ectrodactyly and the type of facial clefting. (For discussion, see text.) DNA binding = DBD; ISO = ISO domain; SAM = SAM domain.
Figure 4
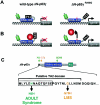
A gain-of-function mutation in ADULT syndrome reveals a second TA domain (TA2) in p63. A and B, Models explaining the gain-of-function effect that the R298Q mutation has on the ΔN-p63γ isotype, which normally does not posses transactivation. The TA2 domain is normally kept in an inactive state, either because of intramolecular interaction (A) or because of binding of another protein (B). This inhibition is proposed to be released in patients with ADULT syndrome because the R298Q mutation abolishes this protein-protein interaction. DNA-BD = DBD; Iso = ISO domain; TA2 = TA2 domain. C, Position of TA2 domain, as determined by Dohn et al. (2001). The TA2 domain consists of 14 amino acids specific to the ΔN isotypes and 12 amino acids common to all p63 isotypes. Interestingly, another ADULT-syndrome mutation and an LMS mutation both give rise to amino acid substitutions within the TA2 domain. DNA binding = DBD; Iso = ISO domain.
Similar articles
- Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63.
McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H. McGrath JA, et al. Hum Mol Genet. 2001 Feb 1;10(3):221-9. doi: 10.1093/hmg/10.3.221. Hum Mol Genet. 2001. PMID: 11159940 - Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans.
van Bokhoven H, McKeon F. van Bokhoven H, et al. Trends Mol Med. 2002 Mar;8(3):133-9. doi: 10.1016/s1471-4914(01)02260-2. Trends Mol Med. 2002. PMID: 11879774 Review. - Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome.
Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Celli J, et al. Cell. 1999 Oct 15;99(2):143-53. doi: 10.1016/s0092-8674(00)81646-3. Cell. 1999. PMID: 10535733 - The p53 family member genes are involved in the Notch signal pathway.
Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. Sasaki Y, et al. J Biol Chem. 2002 Jan 4;277(1):719-24. doi: 10.1074/jbc.M108080200. Epub 2001 Oct 18. J Biol Chem. 2002. PMID: 11641404 - p63.
Little NA, Jochemsen AG. Little NA, et al. Int J Biochem Cell Biol. 2002 Jan;34(1):6-9. doi: 10.1016/s1357-2725(01)00086-3. Int J Biochem Cell Biol. 2002. PMID: 11733180 Review.
Cited by
- ΔNp63α exerts antitumor functions in cervical squamous cell carcinoma.
Zhou Y, Liu H, Wang J, Wang X, Qian L, Xu F, Song W, Wu D, Shen Z, Feng D, Ling B, Xiao W, Shan G, Chen L. Zhou Y, et al. Oncogene. 2020 Jan;39(4):905-921. doi: 10.1038/s41388-019-1033-x. Epub 2019 Oct 1. Oncogene. 2020. PMID: 31576015 - p63/p51-induced onset of keratinocyte differentiation via the c-Jun N-terminal kinase pathway is counteracted by keratinocyte growth factor.
Ogawa E, Okuyama R, Egawa T, Nagoshi H, Obinata M, Tagami H, Ikawa S, Aiba S. Ogawa E, et al. J Biol Chem. 2008 Dec 5;283(49):34241-9. doi: 10.1074/jbc.M804101200. Epub 2008 Oct 10. J Biol Chem. 2008. PMID: 18849344 Free PMC article. - Differential PERP regulation by TP63 mutants provides insight into AEC pathogenesis.
Beaudry VG, Pathak N, Koster MI, Attardi LD. Beaudry VG, et al. Am J Med Genet A. 2009 Sep;149A(9):1952-7. doi: 10.1002/ajmg.a.32760. Am J Med Genet A. 2009. PMID: 19353588 Free PMC article. - Prioritization of neurodevelopmental disease genes by discovery of new mutations.
Hoischen A, Krumm N, Eichler EE. Hoischen A, et al. Nat Neurosci. 2014 Jun;17(6):764-72. doi: 10.1038/nn.3703. Epub 2014 May 27. Nat Neurosci. 2014. PMID: 24866042 Free PMC article. Review. - p63 exerts spatio-temporal control of palatal epithelial cell fate to prevent cleft palate.
Richardson R, Mitchell K, Hammond NL, Mollo MR, Kouwenhoven EN, Wyatt ND, Donaldson IJ, Zeef L, Burgis T, Blance R, van Heeringen SJ, Stunnenberg HG, Zhou H, Missero C, Romano RA, Sinha S, Dixon MJ, Dixon J. Richardson R, et al. PLoS Genet. 2017 Jun 12;13(6):e1006828. doi: 10.1371/journal.pgen.1006828. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28604778 Free PMC article.
References
Electronic-Database Information
- IARC TP53 Mutation Database, http://www.iarc.fr/p53/ (for mutation frequencies in the TP53 gene)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for EEC syndrome [MIM 604292], LADD syndrome [MIM 149730], ADULT syndrome [MIM 103285], LMS [MIM 603543], AEC syndrome [MIM 106260], RHS [MIM 129400], ECP syndrome [MIM 129830], EE syndrome [MIM 129810], SHFM1 [MIM 183600], SHFM2 [MIM 313350], SHFM3 [MIM 600095], and SHFM4 [MIM 605289])
References
- Amiel J, Bougeard G, Francannet C, Raclin V, Munnich A, Lyonnet S, Frebourg T (2001) TP63 gene mutation in ADULT syndrome. Eur J Hum Genet 9:642–645 - PubMed
- Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M (2002) Zebrafish ΔNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell 6:617–627 - PubMed
- Bamberger C, Pollet D, Schmale H (2002) Retinoic acid inhibits downregulation of ΔNp63α expression during terminal differentiation of human primary keratinocytes. J Invest Dermatol 118:133–138 - PubMed
- Bamberger C, Schmale H (2001) Identification and tissue distribution of novel KET/p63 splice variants. FEBS Lett 501:121–126 - PubMed
- Bamshad M, Jorde LB, Carey JC (2000) Getting a LEAD on EEC. Am J Med Genet 90:183–184 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous