Essential and instructive roles of GATA factors in eosinophil development - PubMed (original) (raw)
. 2002 Jun 3;195(11):1379-86.
doi: 10.1084/jem.20020170.
Ritsuko Shimizu, Satoru Takahashi, Mitsujiro Osawa, Shu Takayanagi, Yuko Kato, Masafumi Onodera, Naoko Minegishi, Masayuki Yamamoto, Katashi Fukao, Hideki Taniguchi, Hiromitsu Nakauchi, Atsushi Iwama
Affiliations
- PMID: 12045236
- PMCID: PMC2193540
- DOI: 10.1084/jem.20020170
Essential and instructive roles of GATA factors in eosinophil development
Ryutaro Hirasawa et al. J Exp Med. 2002.
Abstract
GATA transcription factors are major regulators of hematopoietic and immune system. Among GATA factors, GATA-1, GATA-2, and GATA-3 play crucial roles in the development of erythroid cells, hematopoietic stem, and progenitor cells, and T helper type 2 (Th2) cells, respectively. A high level of GATA-1 and GATA-2 expression has been observed in eosinophils, but their roles in eosinophil development remain uncertain both in vitro and in vivo. Here we show that enforced expression of GATA-1 in human primary myeloid progenitor cells completely switches myeloid cell fate into eosinophils. Expression of GATA-1 exclusively promotes development and terminal maturation of eosinophils. Functional domain analyses revealed that the COOH-terminal finger is essential for this capacity while the other domains are dispensable. Importantly, GATA-1-deficient mice failed to develop eosinophil progenitors in the fetal liver. On the other hand, GATA-2 also showed instructive capacity comparable to GATA-1 in vitro and efficiently compensated for GATA-1 deficiency in terms of eosinophil development in vivo, indicating that proper accumulation of GATA factors is critical for eosinophil development. Taken together, our findings establish essential and instructive roles of GATA factors in eosinophil development. GATA-1 and GATA-2 could be novel molecular targets for therapeutic approaches to allergic inflammation.
Figures
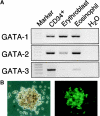
Figure 1.
Expression of GATA family genes in eosinophils. (A) Expression of GATA family genes in human eosinophils. RT-PCR was performed on normalized cDNA templates from cord blood CD34+ cells, glycophorin A+ erythroblasts, and eosinophils obtained by in vitro culture (details in Materials and Methods). Lane H2O represents the negative control without template. (B) Expression of GATA-1 in mouse eosinophils. Eosinophil colonies were generated from bone marrow cells of transgenic mice bearing _IE3.9int_-directed GFP in the presence of IL-5 alone. Bright field microgram (left) and fluorescence microgram (right).
Figure 2.
GATA-1 and GATA-2 promote human eosinophil development. (A) Schematic representation of the retroviral vector, GCsam-IRES-EGFP, harboring wild-type or dominant negative GATA genes linked by an internal ribosome entry site (IRES) to a cDNA encoding EGFP. ψ+, packaging signal; SD, splice donor; SA, splice acceptor; F, Flag-tag; AD, activation domain; NF, NH2-terminal zinc finger; CF, COOH-terminal zinc finger; En, engrailed repression domain. (B) Colony formation of CD34+ cells transduced with wild-type GATA genes. Human CD34+ hematopoietic progenitor cells transduced with empty vector (control), GATA-1 or GATA-2 were cultured in methylcellulose medium. CFU-GM/M, CFU-granulocyte/macrophage and macrophage, BFU-E, erythroid burst-forming unit. Results are shown as mean ± SD per 1,000 cells of three representative experiments. (C) The morphology of an eosinophil colony generated from _GATA-1_–transduced CD34+ progenitor cells. Morphology under a phase-contrast microscope (left). The eosinophil colony was lifted and cytocentrifuged onto a glass slide, then stained by May-Grüenwald Giemsa staining (middle) and Fast green staining (right).
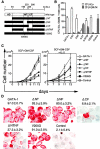
Figure 3.
Effect of GATA genes on eosinophil development. (A) Cell growth and differentiation of CD34+ progenitor cells transduced with either wild-type GATA or dominant negative GATA. Cytokine-dependent growth of transduced cells was evaluated by liquid culture. To evaluate IL-5–dependent cell growth, cells were cultured in the presence of SCF and GM-CSF for the first 5 d. Then, cytokines were replaced to IL-5 alone (middle). Alternatively, preculture media were supplemented with IL-3 in addition to SCF and GM-CSF to facilitate development of eosinophil progenitors (right). Results are shown as mean ± SD of three representative experiments. (B) Flow cytometric profiles of transduced cells cultured for 10 d in the presence of SCF and GM-CSF. The percentages of CD11b+CD14+ macrophages and MBP+ eosinophils are indicated. (C) Eosinophilic features of _GATA_-expressing cells. CD34+ progenitor cells transduced with wild-type GATA genes were cultured for 10 d in the presence of SCF and GM-CSF, then analyzed by May-Grüenwald Giemsa staining (MGG), Fast Green staining (FG), and immunostaining for intracytoplasmic EPO.
Figure 4.
GATA-1 functional domains responsible for eosinophil development. (A) Schematic representation of GATA-1 mutants. (B and C) Growth and differentiation of CD34+ cells transduced with GATA mutants. (B) CD34+ cells transduced with empty vector (control), wild-type GATA-1, or GATA-1 mutants were cultured for 14 d in methylcellulose medium supplemented with SCF and GM-CSF. (C) Cytokine-dependent cell growth was also evaluated by liquid culture as in Fig. 3 A. Results are shown as mean ± SD of three representative experiments. (D) Effects of GATA-1 mutants on eosinophil development. Transduced CD34+ progenitor cells were cultured for 10 d in the presence of SCF and GM-CSF, then analyzed by immunostaining for intracytoplasmic EPO. The percentages of EPO+ eosinophils are indicated as mean ± SD of three representative experiments.
Similar articles
- Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo.
Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Yu C, et al. J Exp Med. 2002 Jun 3;195(11):1387-95. doi: 10.1084/jem.20020656. J Exp Med. 2002. PMID: 12045237 Free PMC article. - Identification of eosinophil lineage-committed progenitors in the murine bone marrow.
Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Iwasaki H, et al. J Exp Med. 2005 Jun 20;201(12):1891-7. doi: 10.1084/jem.20050548. Epub 2005 Jun 13. J Exp Med. 2005. PMID: 15955840 Free PMC article. - Fractionation of mature eosinophils in GATA-reporter transgenic mice.
Kim K, Suzuki N, Ohneda K, Minegishi N, Yamamoto M. Kim K, et al. Tohoku J Exp Med. 2010 Feb;220(2):127-38. doi: 10.1620/tjem.220.127. Tohoku J Exp Med. 2010. PMID: 20139664 - Making eosinophils through subtle shifts in transcription factor expression.
McNagny K, Graf T. McNagny K, et al. J Exp Med. 2002 Jun 3;195(11):F43-7. doi: 10.1084/jem.20020636. J Exp Med. 2002. PMID: 12045250 Free PMC article. Review. No abstract available. - Gene expression regulation and domain function of hematopoietic GATA factors.
Shimizu R, Yamamoto M. Shimizu R, et al. Semin Cell Dev Biol. 2005 Feb;16(1):129-36. doi: 10.1016/j.semcdb.2004.11.001. Epub 2004 Dec 10. Semin Cell Dev Biol. 2005. PMID: 15659347 Review.
Cited by
- SPI1-KLF1/LYL1 axis regulates lineage commitment during endothelial-to-hematopoietic transition from human pluripotent stem cells.
Qu K, Mo S, Huang J, Liu S, Zhang S, Shen J, Yen K. Qu K, et al. iScience. 2024 Jun 28;27(8):110409. doi: 10.1016/j.isci.2024.110409. eCollection 2024 Aug 16. iScience. 2024. PMID: 39108738 Free PMC article. - Eosinophils in obesity and obesity-associated disorders.
Hu Y, Chakarov S. Hu Y, et al. Discov Immunol. 2023 Nov 14;2(1):kyad022. doi: 10.1093/discim/kyad022. eCollection 2023. Discov Immunol. 2023. PMID: 38567054 Free PMC article. Review. - Pathogenic GATA2 genetic variants utilize an obligate enhancer mechanism to distort a multilineage differentiation program.
Katsumura KR, Liu P, Kim JA, Mehta C, Bresnick EH. Katsumura KR, et al. Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2317147121. doi: 10.1073/pnas.2317147121. Epub 2024 Feb 29. Proc Natl Acad Sci U S A. 2024. PMID: 38422019 Free PMC article. - Strain-dependent modifiers exacerbate familial leukemia caused by GATA1-deficiency.
Hirano I, Abe K, Engel JD, Yamamoto M, Shimizu R. Hirano I, et al. Exp Hematol Oncol. 2024 Feb 26;13(1):23. doi: 10.1186/s40164-024-00491-w. Exp Hematol Oncol. 2024. PMID: 38409047 Free PMC article. - Cebp1 and Cebpβ transcriptional axis controls eosinophilopoiesis in zebrafish.
Li G, Sun Y, Kwok I, Yang L, Wen W, Huang P, Wu M, Li J, Huang Z, Liu Z, He S, Peng W, Bei JX, Ginhoux F, Ng LG, Zhang Y. Li G, et al. Nat Commun. 2024 Jan 27;15(1):811. doi: 10.1038/s41467-024-45029-0. Nat Commun. 2024. PMID: 38280871 Free PMC article.
References
- Sanderson, C.J. 1992. Interleukin-5, eosinophils, and disease. Blood. 79:3101–3109. - PubMed
- Gleich, G.J. 2000. Mechanisms of eosinophil-associated inflammation. J. Allergy Clin. Immunol. 105:651–663. - PubMed
- Orkin, S.H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57–64. - PubMed
- Tenen, D.G., R. Hromas, J.D. Licht, and D.-E. Zhang. 1997. Transcription factors, normal myeloid development, and leukemia. Blood. 90:489–519. - PubMed
- Yamaguchi, Y., H. Nishio, K. Kishi, S.J. Ackerman, and T. Suda. 1999. C/EBPβ and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: Implications for C/EBPβ activity in eosinophil gene expression. Blood. 94:1429–1439. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases