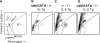Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo - PubMed (original) (raw)
Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo
Channing Yu et al. J Exp Med. 2002.
Abstract
Transcription factor GATA-1 reprograms immature myeloid cells to three different hematopoietic lineages-erythroid cells, megakaryocytes, and eosinophils. GATA-1 is essential for maturation of erythroid and megakaryocytic precursors, as revealed by gene targeting in mice. Here we demonstrate that deletion of a high-affinity GATA-binding site in the GATA-1 promoter, an element presumed to mediate positive autoregulation of GATA-1 expression, leads to selective loss of the eosinophil lineage. These findings suggest that GATA-1 is required for specification of this lineage during hematopoietic development. Mice lacking the ability to produce eosinophils should prove useful in ascertaining the role of eosinophils in a variety of inflammatory or allergic disorders.
Figures
Figure 1.
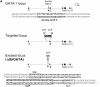
Targeting of the GATA-1 locus. (A, top) The murine GATA-1 locus contains a double GATA-site upstream of the hematopoietic exon IE. Other exons are designated with Roman numerals, including testis exon IT. The double-stranded sequence at the asterisked region is given below the schematic, with nucleotides to be deleted marked in boldface type. (Middle) After targeting, the double GATA-site is deleted, replaced by a floxed PGK-neo cassette. (Bottom) After cre recombinase-mediated excision, the “excised locus” has a minimal sequence consisting primarily of a single loxP site surrounded by two Not I sites. N, Not I site. The double-stranded sequence at the asterisked region is given below the schematic. (B) PCR genotyping of male mice carrying the wt or ΔdblGATA mutant (hem) allele. PCR products before (–) and after (N) digestion with Not I.
Figure 1.
Targeting of the GATA-1 locus. (A, top) The murine GATA-1 locus contains a double GATA-site upstream of the hematopoietic exon IE. Other exons are designated with Roman numerals, including testis exon IT. The double-stranded sequence at the asterisked region is given below the schematic, with nucleotides to be deleted marked in boldface type. (Middle) After targeting, the double GATA-site is deleted, replaced by a floxed PGK-neo cassette. (Bottom) After cre recombinase-mediated excision, the “excised locus” has a minimal sequence consisting primarily of a single loxP site surrounded by two Not I sites. N, Not I site. The double-stranded sequence at the asterisked region is given below the schematic. (B) PCR genotyping of male mice carrying the wt or ΔdblGATA mutant (hem) allele. PCR products before (–) and after (N) digestion with Not I.
Figure 2.
Peripheral blood from a wt male and a ΔdblGATA mutant male mouse. Wright-Giemsa staining of blood smears from a wild-type male (A) and hemizygous ΔdblGATA mutant (B) male. Red cells and platelets appear normal in the ΔdblGATA mutant male. Original magnification: 1,000×.
Figure 3.
Eosinophilia stimulated by an IL-5 transgene is not observed in ΔdblGATA mutant mice. (A) Schematic of cell populations in automated differential analysis of mouse blood. A heterozygous ΔdblGATA mutant female displays a normal-appearing differential plot (B). The IL-5 transgene imparts an increase in the number of eosinophils and large unclassified cells (C) in a male with a wt GATA-1 locus. This increase is not observed in a hemizygous ΔdblGATA mutant male harboring the same transgene (D). (E and F) Wright-Giemsa staining of bone marrow from mice harboring the IL-5 transgene. Numerous eosinophils with bright red granules are evident in a mouse with a wt GATA-1 locus (E) but not in a hemizygous ΔdblGATA mutant male (F). Original magnification: 600×.
Figure 3.
Eosinophilia stimulated by an IL-5 transgene is not observed in ΔdblGATA mutant mice. (A) Schematic of cell populations in automated differential analysis of mouse blood. A heterozygous ΔdblGATA mutant female displays a normal-appearing differential plot (B). The IL-5 transgene imparts an increase in the number of eosinophils and large unclassified cells (C) in a male with a wt GATA-1 locus. This increase is not observed in a hemizygous ΔdblGATA mutant male harboring the same transgene (D). (E and F) Wright-Giemsa staining of bone marrow from mice harboring the IL-5 transgene. Numerous eosinophils with bright red granules are evident in a mouse with a wt GATA-1 locus (E) but not in a hemizygous ΔdblGATA mutant male (F). Original magnification: 600×.
Figure 4.
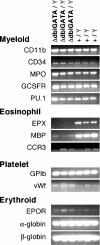
RT-PCR analysis of gene expression in wt and ΔdblGATA mutant male mice. RT-PCR analysis was used to examine gene expression in the bone marrows of hemizygous ΔdblGATA mutant (n = 3) and wt (n = 3) male mice. Control reactions were performed with both wt and mutant samples with all primer pair combinations, either without RT or without cDNA template; in all these cases, no PCR product was generated (data not shown). EPOR, erythropoietin receptor; EPX, eosinophil peroxidase; GCSRFR, granulocyte colony stimulating factor receptor; GPIb, glycoprotein Ib; MBP, major basic protein; MPO, myeloperoxidase; vWf, von Willebrand factor.
Figure 5.
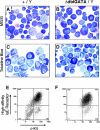
Production of mast cells from wt and ΔdblGATA mutant male mice. May-Grünwald-Giemsa (A and B) and toluidine blue stains (C and D) of cytospin preparations of bone marrow–derived mast cells, from wt (A and C) or ΔdblGATA mutant (B and D) male mice (original magnification: 1,000×). Toluidine blue-positive cells contain dark blue cytoplasmic granules. (E and F) Two-color FACS® analysis of c-kit (x-axis) and high-affinity IgE receptor (y-axis) expression of bone marrow–derived mast cells from wt (E) or ΔdblGATA mutant (F) male mice. Mast cells are observed in both wt and mutant mice.
Similar articles
- A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene.
Nishimura S, Takahashi S, Kuroha T, Suwabe N, Nagasawa T, Trainor C, Yamamoto M. Nishimura S, et al. Mol Cell Biol. 2000 Jan;20(2):713-23. doi: 10.1128/MCB.20.2.713-723.2000. Mol Cell Biol. 2000. PMID: 10611250 Free PMC article. - Essential and instructive roles of GATA factors in eosinophil development.
Hirasawa R, Shimizu R, Takahashi S, Osawa M, Takayanagi S, Kato Y, Onodera M, Minegishi N, Yamamoto M, Fukao K, Taniguchi H, Nakauchi H, Iwama A. Hirasawa R, et al. J Exp Med. 2002 Jun 3;195(11):1379-86. doi: 10.1084/jem.20020170. J Exp Med. 2002. PMID: 12045236 Free PMC article. - Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene.
Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Vyas P, et al. Development. 1999 Jun;126(12):2799-811. doi: 10.1242/dev.126.12.2799. Development. 1999. PMID: 10331989 - [The trend of molecular biology study on eosinophils].
Yamaguchi Y. Yamaguchi Y. Nihon Rinsho. 1993 Mar;51(3):741-6. Nihon Rinsho. 1993. PMID: 8492451 Review. Japanese. - Transcription factor GATA-1 in megakaryocyte development.
Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Orkin SH, et al. Stem Cells. 1998;16 Suppl 2:79-83. doi: 10.1002/stem.5530160710. Stem Cells. 1998. PMID: 11012179 Review.
Cited by
- Macrophage variants in laboratory research: most are well done, but some are RAW.
Herb M, Schatz V, Hadrian K, Hos D, Holoborodko B, Jantsch J, Brigo N. Herb M, et al. Front Cell Infect Microbiol. 2024 Oct 9;14:1457323. doi: 10.3389/fcimb.2024.1457323. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39445217 Free PMC article. Review. - Estrogen signaling suppresses tumor-associated tissue eosinophilia to promote breast tumor growth.
Artham S, Juras PK, Goyal A, Chakraborty P, Byemerwa J, Liu S, Wardell SE, Chakraborty B, Crowder D, Lim F, Strawser CH, Newlin M, Racioppi A, Dent S, Mirminachi B, Roper J, Perou CM, Chang CY, McDonnell DP. Artham S, et al. Sci Adv. 2024 Sep 27;10(39):eadp2442. doi: 10.1126/sciadv.adp2442. Epub 2024 Sep 27. Sci Adv. 2024. PMID: 39331714 Free PMC article. - Eosinophils respond to, but are not essential for control of an acute Salmonella enterica serovar Typhimurium infection in mice.
FitzPatrick RD, Noone JR, Cartwright RA, Gatti DM, Brosschot TP, Lane JM, Jensen EL, Kroker Kimber I, Reynolds LA. FitzPatrick RD, et al. Infect Immun. 2024 Oct 15;92(10):e0032524. doi: 10.1128/iai.00325-24. Epub 2024 Sep 9. Infect Immun. 2024. PMID: 39248486 - Helminth infection driven gastrointestinal hypermotility is independent of eosinophils and mediated by alterations in smooth muscle instead of enteric neurons.
Wang H, Barry K, Zaini A, Coakley G, Moyat M, Daunt CP, Wickramasinghe LC, Azzoni R, Chatzis R, Yumnam B, Camberis M, Le Gros G, Perdijk O, Foong JPP, Bornstein JC, Marsland BJ, Harris NL. Wang H, et al. PLoS Pathog. 2024 Aug 14;20(8):e1011766. doi: 10.1371/journal.ppat.1011766. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39141685 Free PMC article. - Generation and repair of thymic epithelial cells.
Anderson G, Cosway EJ, James KD, Ohigashi I, Takahama Y. Anderson G, et al. J Exp Med. 2024 Oct 7;221(10):e20230894. doi: 10.1084/jem.20230894. Epub 2024 Jul 9. J Exp Med. 2024. PMID: 38980292 Free PMC article. Review.
References
- Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250–1262. - PubMed
- Ito, E., T. Toki, H. Ishihara, H. Ohtani, L. Gu, M. Yokoyama, J.D. Engel, and M. Yamamoto. 1993. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature. 362:466–468. - PubMed
- Martin, D.I., L.I. Zon, G. Mutter, and S.H. Orkin. 1990. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 344:444–447. - PubMed
- Romeo, P.H., M.H. Prandini, V. Joulin, V. Mignotte, M. Prenant, W. Vainchenker, G. Marguerie, and G. Uzan. 1990. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 344:447–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases