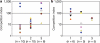Host-induced epidemic spread of the cholera bacterium - PubMed (original) (raw)
Host-induced epidemic spread of the cholera bacterium
D Scott Merrell et al. Nature. 2002.
Abstract
The factors that enhance the transmission of pathogens during epidemic spread are ill defined. Water-borne spread of the diarrhoeal disease cholera occurs rapidly in nature, whereas infection of human volunteers with bacteria grown in vitro is difficult in the absence of stomach acid buffering. It is unclear, however, whether stomach acidity is a principal factor contributing to epidemic spread. Here we report that characterization of Vibrio cholerae from human stools supports a model whereby human colonization creates a hyperinfectious bacterial state that is maintained after dissemination and that may contribute to epidemic spread of cholera. Transcriptional profiling of V. cholerae from stool samples revealed a unique physiological and behavioural state characterized by high expression levels of genes required for nutrient acquisition and motility, and low expression levels of genes required for bacterial chemotaxis.
Figures
Figure 1
Human-shed V. cholerae are hyperinfectious in competition assays in infant mice. a, b, Freshly isolated stool samples from six patients (representing experiments 1, 2 and 3 on the x axes in a and b) were either mixed directly with _in vitro_-grown DSM-V984 (a), or incubated in local pond water for 5 h before mixing (b), and then used for intragastric infection of animals. Data points represent the output ratio of stool-derived V. cholerae to _in vitro_-grown competitor strain after correction for deviations in input ratios. Each data point represents the output from a single animal and symbols and colours are used to facilitate visual discrimination of data values. The geometric mean is indicated by a horizontal bar. n, total number of animals per experiment.
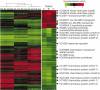
Figure 2
Transcription profile of human-shed V. cholerae. Cluster diagram showing the whole-genome expression profile of V. cholerae recovered from the stools of three ICDDR patients (left panel). Patient samples are labelled A–C and replicate arrays are indicated by the numbers 1–4. The dendrogram shows tight clustering of technical replicates. The right panel is a cluster diagram of genes showing consistent differential regulation from patients A, B and C. Fold changes relative to V. cholerae grown in vitro were calculated using the average values from the quadruplicate arrays shown on the left, and are schematically represented here. Red indicates a minimum twofold increase in expression; green represents a minimum twofold reduction in expression. Representative genes are listed and their relative location indicated by an arrow. See Supplementary Information for a complete list with relative fold changes.
Similar articles
- A prospective cohort study comparing household contact and water Vibrio cholerae isolates in households of cholera patients in rural Bangladesh.
George CM, Hasan K, Monira S, Rahman Z, Saif-Ur-Rahman KM, Rashid MU, Zohura F, Parvin T, Islam Bhuyian MS, Mahmud MT, Li S, Perin J, Morgan C, Mustafiz M, Sack RB, Sack DA, Stine OC, Alam M. George CM, et al. PLoS Negl Trop Dis. 2018 Jul 27;12(7):e0006641. doi: 10.1371/journal.pntd.0006641. eCollection 2018 Jul. PLoS Negl Trop Dis. 2018. PMID: 30052631 Free PMC article. - Cholera stool bacteria repress chemotaxis to increase infectivity.
Butler SM, Nelson EJ, Chowdhury N, Faruque SM, Calderwood SB, Camilli A. Butler SM, et al. Mol Microbiol. 2006 Apr;60(2):417-26. doi: 10.1111/j.1365-2958.2006.05096.x. Mol Microbiol. 2006. PMID: 16573690 Free PMC article. - Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae.
Bourassa L, Camilli A. Bourassa L, et al. Mol Microbiol. 2009 Apr;72(1):124-38. doi: 10.1111/j.1365-2958.2009.06629.x. Epub 2009 Feb 17. Mol Microbiol. 2009. PMID: 19226328 Free PMC article. - Cholera transmission: the host, pathogen and bacteriophage dynamic.
Nelson EJ, Harris JB, Morris JG Jr, Calderwood SB, Camilli A. Nelson EJ, et al. Nat Rev Microbiol. 2009 Oct;7(10):693-702. doi: 10.1038/nrmicro2204. Nat Rev Microbiol. 2009. PMID: 19756008 Free PMC article. Review. - [Factors contributing to preservation of Vibrio cholerae in water reservoirs].
Mikhaĭlova AE, Khaĭtovich AB. Mikhaĭlova AE, et al. Zh Mikrobiol Epidemiol Immunobiol. 2000 Nov-Dec;(6):99-104. Zh Mikrobiol Epidemiol Immunobiol. 2000. PMID: 11210656 Review. Russian.
Cited by
- Quorum regulated latent environmental cells of toxigenic Vibrio cholerae and their role in cholera outbreaks.
Faruque SN, Yamasaki S, Faruque SM. Faruque SN, et al. Gut Pathog. 2024 Sep 29;16(1):52. doi: 10.1186/s13099-024-00647-3. Gut Pathog. 2024. PMID: 39343919 Free PMC article. Review. - Core and accessory genomic traits of Vibrio cholerae O1 drive lineage transmission and disease severity.
Maciel-Guerra A, Babaarslan K, Baker M, Rahman A, Hossain M, Sadique A, Alam J, Uzzaman S, Ferdous Rahman Sarker M, Sultana N, Islam Khan A, Ara Begum Y, Hassan Afrad M, Senin N, Hossain Habib Z, Shirin T, Qadri F, Dottorini T. Maciel-Guerra A, et al. Nat Commun. 2024 Sep 23;15(1):8231. doi: 10.1038/s41467-024-52238-0. Nat Commun. 2024. PMID: 39313510 Free PMC article. - Cholera rages in Africa and the Middle East: A narrative review on challenges and solutions.
Ahmed AK, Sijercic VC, Akhtar MS, Elbayomy A, Marouf MA, Zeleke MS, Sayad R, Abdelshafi A, Laird NJ, El-Mokhtar MA, Ruthig GR, Hetta HF. Ahmed AK, et al. Health Sci Rep. 2024 May 12;7(5):e2013. doi: 10.1002/hsr2.2013. eCollection 2024 May. Health Sci Rep. 2024. PMID: 38742091 Free PMC article. Review. - Case-area targeted interventions during a large-scale cholera epidemic: A prospective cohort study in Northeast Nigeria.
OKeeffe J, Salem-Bango L, Desjardins MR, Lantagne D, Altare C, Kaur G, Heath T, Rangaiya K, Obroh PO, Audu A, Lecuyot B, Zoungrana T, Ihemezue EE, Aye S, Sikder M, Doocy S, Wang Q, Xiao M, Spiegel PB. OKeeffe J, et al. PLoS Med. 2024 May 10;21(5):e1004404. doi: 10.1371/journal.pmed.1004404. eCollection 2024 May. PLoS Med. 2024. PMID: 38728366 Free PMC article. - On or Off: Life-Changing Decisions Made by Vibrio cholerae Under Stress.
Zhou Y, Lee ZL, Zhu J. Zhou Y, et al. Infect Microbes Dis. 2020 Oct 14;2(4):127-135. doi: 10.1097/IM9.0000000000000037. eCollection 2020 Dec. Infect Microbes Dis. 2020. PMID: 38630076 Free PMC article. Review.
References
- Cash RA, et al. Response of man to infection with Vibrio cholerae. I. Clinical, serologic, and bacteriologic responses to a known inoculum. J. Infect. Dis. 1974;129:45–52. - PubMed
- Gitelson S. Gastrectomy, achlorhydria and cholera. Israel J. Med. Sci. 1971;7:663–667. - PubMed
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
- Freter R, Smith HL, Sweeney FJ. Enumeration of cholera vibrios in faecal samples. J. Infect. Dis. 1961;109:31–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical