Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824 - PubMed (original) (raw)
Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824
Latonia M Harris et al. J Bacteriol. 2002 Jul.
Abstract
The Clostridium acetobutylicum ATCC 824 spo0A gene was cloned, and two recombinant strains were generated, an spo0A inactivation strain (SKO1) and an spo0A overexpression strain [824(pMPSOA)]. SKO1 was developed by targeted gene inactivation with a replicative plasmid capable of double-crossover chromosomal integration--a technique never used before with solventogenic clostridia. SKO1 was severely deficient in solvent formation: it produced only 2 mM acetone and 13 mM butanol, compared to the 92 mM acetone and 172 mM butanol produced by the parental strain. After 72 h of growth on solid media, SKO1 formed long filaments of rod-shaped cells that failed to septate. SKO1 cells never achieved the swollen clostridial form typical of the parental strain and did not form endospores. No spo0A transcripts were detected in SKO1, while transcription of two solvent formation operons (aad-ctfA-ctfB and adc; both containing 0A boxes in their promoter regions) was limited. Strain 824(pMSPOA) produced higher butanol concentrations than the control strain [824(pIMP1)] and dramatically elevated spo0A transcript levels and displayed a bimodal pattern of spo0A transcription similar to that of B. subtilis. Microscopic studies indicated that sporulation was both enhanced and accelerated due to spo0A overexpression compared to that of both the 824(pIMP1) and parental strains. Consistent with that, expression of the key solvent formation genes (aad-ctfA-ctfB and adc) and three sporulation-specific genes (spoIIGA, sigE, and sigG) was observed earlier in strain 824(pMSPOA) than in the plasmid control. These data support the hypothesis that Spo0A is a transcriptional regulator that positively controls sporulation and solvent production. Its effect on solvent formation is a balancing act in regulating sporulation versus solvent gene expression: its overexpression apparently tips the balance in favor of accelerated and enhanced sporulation at the expense of overall solvent production.
Figures
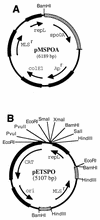
FIG. 1.
Partial restriction maps of plasmids pMSPOA (A) and pETSPO (B). In pETSPO, the spo0A fragments are checkered.
FIG. 2.
Putative crossover events during spo0A gene inactivation. spo0A sequences are checkered, and the MLSr-encoding gene is represented by an open segment. All other sequences are shown as black lines or filled segments.
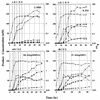
FIG. 3.
Representative fermentation kinetics from bioreactor experiments of strain SKO1 (I), the parental strain (II), strain 824(pMSPOA) (III), and strain 824(pIMP1) (IV). Samples from these experiments were used for the Northern analysis data presented in Fig. 4 and 5. The five sampling points (A, B, C, D, and E) for each experiment are indicated by solid vertical lines. Symbols: ×, _A_600; ▾, butanol concentration; •, acetone concentration; ♦, ethanol concentration; ▿, butyrate concentration; ○, acetate concentration.
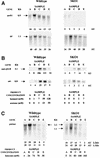
FIG. 4.
Parental (wild-type) and SKO1 strain Northern analyses with the spo0A and thl probes (A), the aad-ctfA/B and adc probes (B), and the ptb-buk probe (C). RNA samples collected during the exponential growth phase (samples A [h 6 and 6.3; _A_600 of 0.57 and 0.47 for the SKO1 and parental strains, respectively] and B [h 10 and 9.5; _A_600 of 3 and 4.7 for the SKO1 and parental strains, respectively]), the transition state (samples C [h 15; _A_600 of 4.6 and 6.7 for the SKO1 and parental strains, respectively]), and the stationary phase (samples D [h 20; _A_600 of 5.6 and 9.5 for the SKO1 and parental strains, respectively] and E [h 25; _A_600 of 5.6 and 9.9 for the SKO1 and parental strains, respectively]) of fermentations (Fig. 3) were electrophoretically resolved and probed with 32P-labeled probes. The blots were electronically imaged with InstantImager Electronic Autoradiography, and the total radioactive counts were measured after 10 min of imaging for spo0A, aad-ctfA-ctfB, and adc and 20 min of imaging for thl and ptb-buk. The mRNA levels are reported in arbitrary units (AU), which are the total counts divided by 1,000.
FIG. 5.
Northern analysis of strains 824(pIMP1) and 824(pMSPOA) with the spo0A and thl probes (A), the ptb-buk probe (B), the aad-ctfA-ctfB and adc probes (C), and the sigE-sigG probe (D). RNA samples collected during the exponential growth phase {samples A [h 6.3 and 6.5; _A_600 of 0.74 and 0.54 for the 824(pMSPOA) and 824(pIMP1) strains, respectively] and B [h 9.5 and 9.5; _A_600 of 3.4 and 1.8 for the 824(pMSPOA) and 824(pIMP1) strains, respectively]}, the transition state {samples C [h 14 and 13.5; _A_600 of 8.2 and 4.9 for the 824(pMSPOA) and 824(pIMP1) strains, respectively]}, and the stationary phase {samples D [h 19 and 18.8; _A_600 of 9.4 and 6.2 for the 824(pMSPOA) and 824(pIMP1) strains, respectively] and E [h 24.3 and 24.8; _A_600 of 9.6 and 5.8, for the 824(pMSPOA) and 824(pIMP1) strains, respectively]} of fermentations (Fig. 3) were electrophoretically resolved and probed with 32P-labeled probes. The blots were electronically imaged with InstantImager Electronic Autoradiography, and the total radioactive counts were measured after 10 min of imaging for spo0A, aad-ctfA-ctfB, and adc and 20 min of imaging for thl, ptb-buk, and sigE-sigG. The mRNA levels are reported in arbitrary units (AU), which are the total counts divided by 1,000.
Similar articles
- Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress.
Alsaker KV, Spitzer TR, Papoutsakis ET. Alsaker KV, et al. J Bacteriol. 2004 Apr;186(7):1959-71. doi: 10.1128/JB.186.7.1959-1971.2004. J Bacteriol. 2004. PMID: 15028679 Free PMC article. - Comparison of expression of key sporulation, solventogenic and acetogenic genes in C. beijerinckii NRRL B-598 and its mutant strain overexpressing spo0A.
Kolek J, Diallo M, Vasylkivska M, Branska B, Sedlar K, López-Contreras AM, Patakova P. Kolek J, et al. Appl Microbiol Biotechnol. 2017 Nov;101(22):8279-8291. doi: 10.1007/s00253-017-8555-3. Epub 2017 Oct 8. Appl Microbiol Biotechnol. 2017. PMID: 28990140 - DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5.
Tomas CA, Alsaker KV, Bonarius HP, Hendriksen WT, Yang H, Beamish JA, Paredes CJ, Papoutsakis ET. Tomas CA, et al. J Bacteriol. 2003 Aug;185(15):4539-47. doi: 10.1128/JB.185.15.4539-4547.2003. J Bacteriol. 2003. PMID: 12867463 Free PMC article. - Transcriptional regulation of solventogenesis in Clostridium acetobutylicum.
Dürre P, Böhringer M, Nakotte S, Schaffer S, Thormann K, Zickner B. Dürre P, et al. J Mol Microbiol Biotechnol. 2002 May;4(3):295-300. J Mol Microbiol Biotechnol. 2002. PMID: 11931561 Review. - The Clostridium sporulation programs: diversity and preservation of endospore differentiation.
Al-Hinai MA, Jones SW, Papoutsakis ET. Al-Hinai MA, et al. Microbiol Mol Biol Rev. 2015 Mar;79(1):19-37. doi: 10.1128/MMBR.00025-14. Microbiol Mol Biol Rev. 2015. PMID: 25631287 Free PMC article. Review.
Cited by
- Production of propionate using metabolically engineered strains of Clostridium saccharoperbutylacetonicum.
Baur T, Wentzel A, Dürre P. Baur T, et al. Appl Microbiol Biotechnol. 2022 Nov;106(22):7547-7562. doi: 10.1007/s00253-022-12210-8. Epub 2022 Oct 25. Appl Microbiol Biotechnol. 2022. PMID: 36282302 Free PMC article. - A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum.
Zhang J, Liu YJ, Cui GZ, Cui Q. Zhang J, et al. Biotechnol Biofuels. 2015 Mar 4;8:36. doi: 10.1186/s13068-015-0214-2. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 25763107 Free PMC article. - Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum.
Tummala SB, Welker NE, Papoutsakis ET. Tummala SB, et al. J Bacteriol. 2003 Mar;185(6):1923-34. doi: 10.1128/JB.185.6.1923-1934.2003. J Bacteriol. 2003. PMID: 12618456 Free PMC article. - Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum.
Alsaker KV, Papoutsakis ET. Alsaker KV, et al. J Bacteriol. 2005 Oct;187(20):7103-18. doi: 10.1128/JB.187.20.7103-7118.2005. J Bacteriol. 2005. PMID: 16199581 Free PMC article. - Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant.
Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, Hogsett DA, Caiazza NC. Tripathi SA, et al. Appl Environ Microbiol. 2010 Oct;76(19):6591-9. doi: 10.1128/AEM.01484-10. Epub 2010 Aug 6. Appl Environ Microbiol. 2010. PMID: 20693441 Free PMC article.
References
- Bahl, H., H. Muller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:341-348. - PubMed
- Brown, D. P., L. Ganova-Raeva, B. D. Green, S. R. Wilkinson, M. Young, and P. Youngman. 1994. Characterization of spo0A homologues in diverse Bacillus and Clostridium species reveals regions of high conservation within the effector domain. Mol. Microbiol. 14:411-426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases