Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis - PubMed (original) (raw)
Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis
Wen-Hui Shen et al. Mol Biol Cell. 2002 Jun.
Abstract
The SCF (for SKP1, Cullin/CDC53, F-box protein) ubiquitin ligase targets a number of cell cycle regulators, transcription factors, and other proteins for degradation in yeast and mammalian cells. Recent genetic studies demonstrate that plant F-box proteins are involved in auxin responses, jasmonate signaling, flower morphogenesis, photocontrol of circadian clocks, and leaf senescence, implying a large spectrum of functions for the SCF pathway in plant development. Here, we present a molecular and functional characterization of plant cullins. The Arabidopsis genome contains 11 cullin-related genes. Complementation assays revealed that AtCUL1 but not AtCUL4 can functionally complement the yeast cdc53 mutant. Arabidopsis mutants containing transfer DNA (T-DNA) insertions in the AtCUL1 gene were shown to display an arrest in early embryogenesis. Consistently, both the transcript and the protein of the AtCUL1 gene were found to accumulate in embryos. The AtCUL1 protein localized mainly in the nucleus but also weakly in the cytoplasm during interphase and colocalized with the mitotic spindle in metaphase. Our results demonstrate a critical role for the SCF ubiquitin ligase in Arabidopsis embryogenesis.
Figures
Figure 1
Sequence analysis of Arabidopsis_cullin-related proteins. (A) Phylogenetic tree of the_Arabidopsis proteins (bold letters), together with cullins and APC2 of Saccharomyces cerevisiae, C. elegans, and Homo sapiens, was established by use of ClustalW and TreeViewPPC programs. DDBJ/EMBL/GenBank accession numbers: Q12018 for ScCDC53, P53202 for ScCUL-B, NP_012488 for ScCUL-C, NP_013228 for ScAPC2, Q17389 for CeCUL-1, Q17390 for CeCUL-2, Q17391 for CeCUL-3, Q17392 for CeCUL-4, Q23639 for CeCUL-5, Q21346 for CeCUL-6, AAF99984 for CeAPC2, NP_003583 for HsCUL1, NP_003582 for HsCUL2, NP_003581 for HsCUL3, NP_003580 for HsCUL4A, AAK16812 for HsCUL4B, AAK07472 for HsCUL5, NP_037498 for HsAPC2, AAK76704 for At4g02570 (AtCUL1, this work), NP_175007 for At1g43140, NP_171797 for At1g02980, NP_176189 for At1g59800, NP_176188 for At1g59790, NP_174005 for At1g26830, NP_177125 for At1g69670, AJ318018 for At5g46210 (AtCUL4, this work), NP_178543 for At2g04660, NP_192947 for At4g12100, and NP_190275 for At3g46910. (B) Sequence alignment of AtCUL1 (At4g02570) and its most closely related Arabidopsis proteins was performed by use of ClustalX. Numbers refer to amino acid positions in the corresponding proteins. Consensus symbols on top of the alignment: * for the identical or conserved residues in all sequences; : and . for the conserved and semiconserved substitutions, respectively. The RBX1/ROC1 binding domain and the RUB1/NEDD8 conjugation site are indicated by a line and an arrow, respectively.
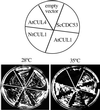
Figure 2
Complementation of the yeast cdc53_mutant by plant cullins. The yeast temperature-sensitive mutant_cdc53 ts was transformed with the empty vector or the vectors expressing CDC53, AtCUL1, NtCUL1, and AtCUL4, respectively. Individual transformants were plated on selective media and grown either at permissive (28°C) or at restrictive (35°C) temperature. Photographs were taken after 4 d.
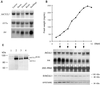
Figure 3
Northern and Western blot analyses of_AtCUL1_ expression. (A) Total RNA was isolated from different organs of Arabidopsis plants, and Northern analysis was performed by successive hybridizations with different probes, as indicated. EFTu: elongation factor EF-1α; H4: histone H4. (B) Samples were taken at different days of subculture from an_Arabidopsis_ cell suspension culture and used for fresh weight measurement and for RNA and protein analysis. Northern analysis was performed by successive hybridizations with the indicated probes. Western blots were performed with the antibodies against AtCUL1 (@AtCUL1) and the conserved CDK kinase motif PSTAIRE (@PSTAIRE). (C) Total proteins prepared from tobacco BY2 cells (lane 1), transgenic BY2 cells expressing 10×his-tagged AtCUL1 (lane 2), and 2-week-old_Arabidopsis_ seedling of wild-type Wassilewskija genotype (lane 3) and Km-resistant_atcul1–1_ +/− (see Table 1) genotype (lane 4) were Western blotted with @AtCUL1. The asterisk indicates an aspecific protein band, which cannot be competed by the AtCUL1 peptide (data not shown).
Figure 4
Vectors used for plant transformation. (A) Schematic representation of the genes inserted into different vectors. Arrows with lines represent the different promoter regions and open boxes the coding sequence of different proteins. (B) Oligonucleotides used in PCR amplification for plant vector construction. Nucleotides corresponding to the AtCUL1 sequence are specified by capital letters. Restriction enzyme sites used in cloning are underlined.
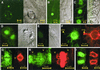
Figure 5
Subcellular localization of AtCUL1. (A) Leaf epidermal pavement and stomata cells of a transgenic tobacco plant expression GFP-AtCUL1. (B) Root cortex cells of a transgenic tobacco plant expression GFP-AtCUL1. (C) Transgenic TBY2 cells expressing GFP-AtCUL1. (D) Arabidopsis suspension culture cells showing immunofluorescence after incubation with @AtCUL1 as primary antibody and the Alexa 488-conjugated goat anti-rabbit IgG (Molecular Probes) as secondary antibody. (E) Transgenic TBY2 cells expressing 10×His-tagged AtCUL1 under the control of CaMV 35S promoter were used for immunolocalization with rabbit @AtCUL1 preimmune serum. (F to I) Transgenic TBY2 cells expressing 10×His-tagged AtCUL1 used for coimmunolocalization studies using as primary antibodies the rabbit @AtCUL1 and the mouse @α-tubulin, together with their corresponding secondary antibodies (the Alexa 488–conjugated goat anti-rabbit IgG and the Alexa 568–conjugated goat antimouse IgG, respectively). The fluorescence of Alexa 488 (green, representing AtCUL1) and Alexa 568 (red, representing α-tubulin) was visualized in preprophase (F), metaphase (G), telophase (H), and early interphase (I) cells. > indicates the chromosome position. PPB, preprophase band.
Figure 6
Schematic representation of the T-DNA insertions in the AtCUL1 gene. The comparison between the genomic and cDNA sequences of the AtCUL1 revealed that the coding region of the gene consists of 19 exons (black boxes) separated by 18 introns. The junctions between AtCUL1 and T-DNA are detailed by the representation of the AtCUL1 exon sequence in triple-nucleotide codon format, intron in italics, and those of unknown origin (corresponding neither to _AtCUL1_nor to T-DNA) in underlined letters. LB and RB indicate the orientation of the left and right borders of the T-DNA, respectively.
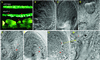
Figure 7
Phenotypes of atcul1 mutants. (A) Open siliques from self-pollinated wild-type and_atcul1–1_ mutant plants. Arrows indicate the developmental arrested siblings. (B–H) Differential interference contrast images of cleared ovules from self-pollinated_atcul1–1_ mutant plants. (B–D) Embryogenesis of developing siblings (containing wild-type or heterozygous zygotes and endosperm) at preglobular (B), globular (C), and heart (D) stages. (E–H) Ovules of arrested siblings from siliques of globular to early heart age. The arrested zygote (arrow) and endosperm (asterisk) cells contain 1 or 2 nuclei each.
Figure 8
Expression of AtCUL1 in embryos. (A) Negative control of in situ hybridization with_AtCUL1_ sense probe in a flower section. (B) A flower section probed with the AtCUL1 antisense probe showing strong staining in ovules of a silique. (C) Close-up of an ovule from (B) showing staining of the embryo. (D) Negative control of immunostaining with the preimmune serum of an ovule containing globular embryo. (E) Immunostaining of an ovule containing globular embryo with @AtCUL1 antibodies, showing strong staining of the embryo. (F) Close-up of the stained embryo from (F) showing strong staining in the nuclei.
Similar articles
- Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response.
Ren C, Pan J, Peng W, Genschik P, Hobbie L, Hellmann H, Estelle M, Gao B, Peng J, Sun C, Xie D. Ren C, et al. Plant J. 2005 May;42(4):514-24. doi: 10.1111/j.1365-313X.2005.02394.x. Plant J. 2005. PMID: 15860010 - The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis.
Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. Xu L, et al. Plant Cell. 2002 Aug;14(8):1919-35. doi: 10.1105/tpc.003368. Plant Cell. 2002. PMID: 12172031 Free PMC article. - SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase.
Farrás R, Ferrando A, Jásik J, Kleinow T, Okrész L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C. Farrás R, et al. EMBO J. 2001 Jun 1;20(11):2742-56. doi: 10.1093/emboj/20.11.2742. EMBO J. 2001. PMID: 11387208 Free PMC article. - AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response.
del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. del Pozo JC, et al. Plant Cell. 2002 Feb;14(2):421-33. doi: 10.1105/tpc.010282. Plant Cell. 2002. PMID: 11884684 Free PMC article. - A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin.
Willems AR, Schwab M, Tyers M. Willems AR, et al. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):133-70. doi: 10.1016/j.bbamcr.2004.09.027. Biochim Biophys Acta. 2004. PMID: 15571813 Review.
Cited by
- Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway.
Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Derrien B, et al. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15942-6. doi: 10.1073/pnas.1209487109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23019378 Free PMC article. - The Arabidopsis F-box protein FBW2 targets AGO1 for degradation to prevent spurious loading of illegitimate small RNA.
Hacquard T, Clavel M, Baldrich P, Lechner E, Pérez-Salamó I, Schepetilnikov M, Derrien B, Dubois M, Hammann P, Kuhn L, Brun D, Bouteiller N, Baumberger N, Vaucheret H, Meyers BC, Genschik P. Hacquard T, et al. Cell Rep. 2022 Apr 12;39(2):110671. doi: 10.1016/j.celrep.2022.110671. Cell Rep. 2022. PMID: 35417704 Free PMC article. - Characterization and comparative expression analysis of CUL1 genes in rice.
Kim SH, Woo OG, Jang H, Lee JH. Kim SH, et al. Genes Genomics. 2018 Mar;40(3):233-241. doi: 10.1007/s13258-017-0622-8. Epub 2017 Oct 19. Genes Genomics. 2018. PMID: 29892794 - Mass Spectrometric Analyses Reveal a Central Role for Ubiquitylation in Remodeling the Arabidopsis Proteome during Photomorphogenesis.
Aguilar-Hernández V, Kim DY, Stankey RJ, Scalf M, Smith LM, Vierstra RD. Aguilar-Hernández V, et al. Mol Plant. 2017 Jun 5;10(6):846-865. doi: 10.1016/j.molp.2017.04.008. Epub 2017 Apr 28. Mol Plant. 2017. PMID: 28461270 Free PMC article. - Effect of Paternal Genome Excess on the Developmental and Gene Expression Profiles of Polyspermic Zygotes in Rice.
Deushi R, Toda E, Koshimizu S, Yano K, Okamoto T. Deushi R, et al. Plants (Basel). 2021 Jan 28;10(2):255. doi: 10.3390/plants10020255. Plants (Basel). 2021. PMID: 33525652 Free PMC article.
References
- Andrade MA, Gonzalez-Guzman M, Serrano R, Rodriguez PL. A combination of the F-box motif and kelch repeats defines a large Arabidopsisfamily of F-box proteins. Plant Mol Biol. 2001;46:603–614. - PubMed
- Arabidopsis genome initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. - PubMed
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199.
- Berger F. Endosperm development. Curr Opin Plant Biol. 1999;2:28–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases