A monomeric red fluorescent protein - PubMed (original) (raw)
A monomeric red fluorescent protein
Robert E Campbell et al. Proc Natl Acad Sci U S A. 2002.
Abstract
All coelenterate fluorescent proteins cloned to date display some form of quaternary structure, including the weak tendency of Aequorea green fluorescent protein (GFP) to dimerize, the obligate dimerization of Renilla GFP, and the obligate tetramerization of the red fluorescent protein from Discosoma (DsRed). Although the weak dimerization of Aequorea GFP has not impeded its acceptance as an indispensable tool of cell biology, the obligate tetramerization of DsRed has greatly hindered its use as a genetically encoded fusion tag. We present here the stepwise evolution of DsRed to a dimer and then either to a genetic fusion of two copies of the protein, i.e., a tandem dimer, or to a true monomer designated mRFP1 (monomeric red fluorescent protein). Each subunit interface was disrupted by insertion of arginines, which initially crippled the resulting protein, but red fluorescence could be rescued by random and directed mutagenesis totaling 17 substitutions in the dimer and 33 in mRFP1. Fusions of the gap junction protein connexin43 to mRFP1 formed fully functional junctions, whereas analogous fusions to the tetramer and dimer failed. Although mRFP1 has somewhat lower extinction coefficient, quantum yield, and photostability than DsRed, mRFP1 matures >10 times faster, so that it shows similar brightness in living cells. In addition, the excitation and emission peaks of mRFP1, 584 and 607 nm, are approximately 25 nm red-shifted from DsRed, which should confer greater tissue penetration and spectral separation from autofluorescence and other fluorescent proteins.
Figures
Figure 1
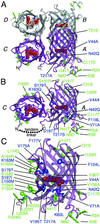
Graphical representation of the tetramer, dimer, and monomer of DsRed based on the x-ray crystal structure of DsRed (21). Residues 1–5 were not observed in the crystal structure (Protein Data Bank identification 1G7K) but have been arbitrarily appended for the sake of representation. The DsRed chromophore is represented in red, and the four chains of the dimer are labeled following the convention of Yarbrough et al. (21). (A) The tetramer of DsRed with all residues mutated in T1 indicated in green for external residues and blue for those internal to the β-barrel. (B) The AC dimer of DsRed with all mutations present in dimer2 represented as in A and the intersubunit linker present in tdimer2(12) shown as a dotted line. (C) The monomer of DsRed with all mutations present in mRFP1 represented as in A. This figure was produced with
molscript
(27).
Figure 2
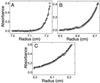
Analytical ultracentrifugation analysis of DsRed, dimer2, and mRFP0.5a. The equilibrium radial absorbance profiles at 20,000 rpm were modeled with a theoretical curve that allowed only the molecular weight to vary. (A) The DsRed absorbance profile was best fit with an apparent molecular mass of 120 kDa, consistent with a tetramer. (B) The dimer2 absorbance profile was best fit with an apparent mass weight of 60 kDa, consistent with a dimer. (C) The mRFP0.5a absorbance profile was best fit with an apparent molecular mass of 32 kDa, consistent with a monomer containing an N-terminal polyhistidine affinity tag.
Figure 3
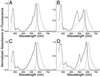
Fluorescence and absorption spectra of DsRed (A), T1 (B), dimer2 and tdimer2(12) (C), and mRFP1 (D). The absorbance spectrum is shown with a solid line, the excitation with a dotted line, and the emission with a dashed line.
Figure 4
Maturation of red fluorescence for DsRed, T1, dimer2, tdimer2(12), and mRFP1. Log-phase cultures of E. coli expressing the construct of interest were rapidly purified at 4°C, and beginning at 2 h postharvest their maturation at 37°C was monitored. The initial decrease in mRFP1 fluorescence is attributed to a slight quenching on warming from 4°C to 37°C.
Figure 5
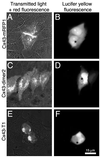
HeLa cells expressing Cx43 fused with T1, dimer2, or mRFP1. (A, C, and E) Images were acquired with excitation at 568 nm (55 nm bandwidth) and emission at 653 nm (95 nm bandwidth) with additional transmitted light. Lucifer yellow fluorescence (B, D, and F) was acquired with excitation at 425 nm (45 nm bandpass) and emission at 535 nm (55 nm bandpass). (A) Two contacting cells transfected with Cx43-mRFP1 and connected by a single large gap junction. (B) One cell is microinjected with lucifer yellow at the point indicated by * and the dye quickly passes (1–2 s) to the adjacent cell. (C) Four neighboring cells transfected with Cx43-dimer2. The bright line between the two rightmost cells is the result of having two fluorescent membranes in contact and is not a gap junction. (D) As was observed about one-third of the time, microinjected dye is slowly passed to an adjacent cell. (E) Two adjacent cells transfected with Cx43-T1 and displaying the typical perinuclear localized aggregation. (F) No dye passed between neighboring cells.
Similar articles
- Genetic and spectrally distinct in vivo imaging: embryonic stem cells and mice with widespread expression of a monomeric red fluorescent protein.
Long JZ, Lackan CS, Hadjantonakis AK. Long JZ, et al. BMC Biotechnol. 2005 Jul 4;5:20. doi: 10.1186/1472-6750-5-20. BMC Biotechnol. 2005. PMID: 15996270 Free PMC article. - Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral.
Baird GS, Zacharias DA, Tsien RY. Baird GS, et al. Proc Natl Acad Sci U S A. 2000 Oct 24;97(22):11984-9. doi: 10.1073/pnas.97.22.11984. Proc Natl Acad Sci U S A. 2000. PMID: 11050229 Free PMC article. - Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer.
Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Mizuno H, et al. Biochemistry. 2001 Feb 27;40(8):2502-10. doi: 10.1021/bi002263b. Biochemistry. 2001. PMID: 11327872 - Green fluorescent protein.
Chalfie M. Chalfie M. Photochem Photobiol. 1995 Oct;62(4):651-6. doi: 10.1111/j.1751-1097.1995.tb08712.x. Photochem Photobiol. 1995. PMID: 7480149 Review. - Anthozoa red fluorescent protein in biosensing.
Shrestha S, Deo SK. Shrestha S, et al. Anal Bioanal Chem. 2006 Oct;386(3):515-24. doi: 10.1007/s00216-006-0652-6. Epub 2006 Aug 1. Anal Bioanal Chem. 2006. PMID: 16924380 Review.
Cited by
- Dynein Separately Partners with NDE1 and Dynactin To Orchestrate T Cell Focused Secretion.
Nath S, Christian L, Tan SY, Ki S, Ehrlich LI, Poenie M. Nath S, et al. J Immunol. 2016 Sep 15;197(6):2090-101. doi: 10.4049/jimmunol.1600180. Epub 2016 Aug 17. J Immunol. 2016. PMID: 27534551 Free PMC article. - Imaging flow cytometry analysis of intracellular pathogens.
Haridas V, Ranjbar S, Vorobjev IA, Goldfeld AE, Barteneva NS. Haridas V, et al. Methods. 2017 Jan 1;112:91-104. doi: 10.1016/j.ymeth.2016.09.007. Epub 2016 Sep 15. Methods. 2017. PMID: 27642004 Free PMC article. Review. - Versatile toolbox for high throughput biochemical and functional studies with fluorescent fusion proteins.
Pichler G, Jack A, Wolf P, Hake SB. Pichler G, et al. PLoS One. 2012;7(5):e36967. doi: 10.1371/journal.pone.0036967. Epub 2012 May 11. PLoS One. 2012. PMID: 22606318 Free PMC article. - Erb-b2 Receptor Tyrosine Kinase 2 (ERBB2) Promotes ATG12-Dependent Autophagy Contributing to Treatment Resistance of Breast Cancer Cells.
Chen Y, Wang R, Huang S, Henson ES, Bi J, Gibson SB. Chen Y, et al. Cancers (Basel). 2021 Mar 2;13(5):1038. doi: 10.3390/cancers13051038. Cancers (Basel). 2021. PMID: 33801244 Free PMC article. - Unilateral Cleavage Furrows in Multinucleate Cells.
Bindl J, Molnar ES, Ecke M, Prassler J, Müller-Taubenberger A, Gerisch G. Bindl J, et al. Cells. 2020 Jun 18;9(6):1493. doi: 10.3390/cells9061493. Cells. 2020. PMID: 32570994 Free PMC article.
References
- Matz M V, Fradkov A F, Labas Y A, Savitsky A P, Zaraisky A G, Markelov M L, Lukyanov S A. Nat Biotechnol. 1999;17:969–973. - PubMed
- Tsien R Y. Annu Rev Biochem. 1998;67:509–544. - PubMed
- Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Biochemistry. 2001;40:2502–2510. - PubMed
- Verkhusha V V, Otsuna H, Awasaki T, Oda H, Tsukita S, Ito K. J Biol Chem. 2001;276:29621–29624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM 62114/GM/NIGMS NIH HHS/United States
- U54 GM062114/GM/NIGMS NIH HHS/United States
- 2P30 CA 23100-18/CA/NCI NIH HHS/United States
- NS 27177/NS/NINDS NIH HHS/United States
- R01 NS027177/NS/NINDS NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R37 NS027177/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous