Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats - PubMed (original) (raw)
Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats
Richard Bayliss et al. EMBO J. 2002.
Abstract
Interactions with nucleoporins containing FxFG-repeat cores are crucial for the nuclear import of RanGDP mediated by nuclear transport factor 2 (NTF2). We describe here the 1.9 A resolution crystal structure of yeast NTF2-N77Y bound to a FxFG-nucleoporin core, which provides a basis for understanding this interaction and its role in nuclear trafficking. The two identical FxFG binding sites on the dimeric molecule are formed by residues from each chain of NTF2. Engineered mutants at the interaction interface reduce the binding of NTF2 to nuclear pores and cause reduced growth rates and Ran mislocalization when substituted for the wild-type protein in yeast. Comparison with the crystal structure of FG-nucleoporin cores bound to importin-beta and TAP/p15 identified a number of common features of their binding sites. The structure of the binding interfaces on these transport factors provides a rationale for the specificity of their interactions with nucleoporins that, combined with their weak binding constants, facilitates rapid translocation through NPCs during nuclear trafficking.
Figures
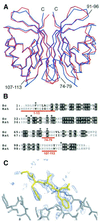
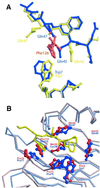
Fig. 1. Structural differences between yeast and rat NTF2. (A) Superposed Cα traces of rNTF2 (blue) and yNTF2-N77Y (red) showing the overall similarity between the two structures. Only the N-terminus (N), C-terminus (C) and the loop containing residues 91–96 differ between all four yNTF2-N77Y chains. (B) Structure-based sequence alignment of yNTF2 (Sc) and rNTF2 (rat). Conserved residues are shown with a black background and the variable regions are marked in red. (C) _F_o – _F_c omit electron density map (blue, contoured at 2σ), generated after simulated annealing of structure after subtraction of the FxFG peptide, showing a tube of density on the surface of yNTF2 (grey) around the FxFG peptide (yellow).
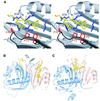
Fig. 2. FxFG-repeat cores bind to a hydrophobic depression at the dimer interface of yNTF2. (A) Stereo drawing showing how the FxFG peptide (chain E, yellow) binds to residues from both chains of the NTF2 dimer (chain A, red and chain B, blue). (B) The relative disposition of the two FxFG cores bound to the NTF2 dimer. The Tyr77 side chains are also shown in grey and three residues (Leu8, Phe12 and Tyr112) that may be involved in a tentative secondary site are in green. (C) The TAP/p15 dimer with bound FG peptide (Fribourg et al., 2001) in the same orientation, illustrating the different locations of the FG-nucleoporin binding sites on each molecule.
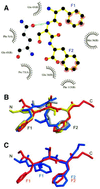
Fig. 3. FxFG–yNTF2 interface. (A) Schematic diagram of molecular contacts between FxFG-repeat core and yNTF2 dimer, adapted from the LIGPLOT output (Wallace et al., 1995). The FxFG core (yellow bonds between black C atoms, red O atoms and blue N atoms) contacts yNTF2 residues (black circles, flashes) primarily by its Phe-ring atoms (red flashes). A putative H-bond (green) is formed between Gln45 (grey bonds) and the FxFG main chain. (B) The backbone conformation of chains E (red) and G (yellow) are different from those seen with chains F (blue) and H (grey), but significantly the Phe rings of all four chains are in approximately the same position. (C) Chains E (red) and G most closely resemble the conformation of FxFG cores bound to importin-β (blue; Bayliss et al., 2000b).
Fig. 4. Surface representations comparing the binding of FG peptides to different transport factors. (A) yNTF2; (B) TAP/p15 (Fribourg et al., 2001); (C) importin-β (Bayliss et al., 2000b). In each case the Phe rings bind to hydrophobic depressions on the transport factor surface, but the overall structure of each site is different.
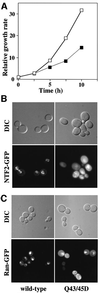
Fig. 5. Q43/45D yNTF2 shows reduced cell growth rate and mislocalization of NTF2 and Ran. (A) When the Q43/45D mutant was expressed in yeast as the only functional copy of NTF2 (filled squares), these cells grew more slowly than wild-type cells (open squares). (B) Localization of GFP fusions to NTF2. Wild-type NTF2 showed primarily the typical punctate nuclear envelope staining characteristic of its binding to NPCs (see Quimby et al., 2001), whereas the Q43/45D mutant showed diffuse staining throughout the cells. (C) With wild-type NTF2, Ran–GFP was localized to the nucleus, whereas with Q43/45D-NTF2 a considerable proportion of the Ran–GFP was mislocalized to the cytoplasm.
Fig. 6. Putative FxFG binding site on rNTF2. (A) Comparison of FxFG binding pocket in wild-type (blue) and D92N/D94N crystal forms (yellow). Crystal lattice interactions in the structure of the D92N/D94N rNTF2 mutant (yellow) include the insertion of a Phe side chain (red) between Gln45 and Gln47. Rotamers adopted by Gln45 and Gln47 in the D92N/D94N structure (yellow) are similar to those found in the yNTF2-N77Y structure, but different from wild-type rNTF2 (blue). In addition, Trp7 moves closer to the inserted Phe aromatic ring. (B) FxFG core (yellow) fits on to the surface of rNTF2 structure (blue), which superposes closely with yNTF2 (red). Rotamers of rNTF2 Gln45 and Gln47 vary between rNTF2 structures. Those shown in the figure are from the structure of rNTF2 bound to RanGDP, which closely match those observed in crystals of D92N/D94N rNTF2, rNTF2-W7A or yNTF2-N77Y.
Similar articles
- Solution NMR study of the interaction between NTF2 and nucleoporin FxFG repeats.
Morrison J, Yang JC, Stewart M, Neuhaus D. Morrison J, et al. J Mol Biol. 2003 Oct 24;333(3):587-603. doi: 10.1016/j.jmb.2003.08.050. J Mol Biol. 2003. PMID: 14556747 - Functional analysis of the hydrophobic patch on nuclear transport factor 2 involved in interactions with the nuclear pore in vivo.
Quimby BB, Leung SW, Bayliss R, Harreman MT, Thirumala G, Stewart M, Corbett AH. Quimby BB, et al. J Biol Chem. 2001 Oct 19;276(42):38820-9. doi: 10.1074/jbc.M105054200. Epub 2001 Aug 6. J Biol Chem. 2001. PMID: 11489893 - Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran.
Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy AJ, Gerace L, Silver PA, Stewart M. Clarkson WD, et al. J Mol Biol. 1997 Oct 10;272(5):716-30. doi: 10.1006/jmbi.1997.1255. J Mol Biol. 1997. PMID: 9368653 - Insights into the molecular mechanism of nuclear trafficking using nuclear transport factor 2 (NTF2).
Stewart M. Stewart M. Cell Struct Funct. 2000 Aug;25(4):217-25. doi: 10.1247/csf.25.217. Cell Struct Funct. 2000. PMID: 11129791 Review. - Fifty years of nuclear pores and nucleocytoplasmic transport studies: multiple tools revealing complex rules.
Floch AG, Palancade B, Doye V. Floch AG, et al. Methods Cell Biol. 2014;122:1-40. doi: 10.1016/B978-0-12-417160-2.00001-1. Methods Cell Biol. 2014. PMID: 24857723 Review.
Cited by
- A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins.
Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, Gopinathan A, Lau EY, Colvin ME, Uversky VN, Rexach MF. Yamada J, et al. Mol Cell Proteomics. 2010 Oct;9(10):2205-24. doi: 10.1074/mcp.M000035-MCP201. Epub 2010 Apr 5. Mol Cell Proteomics. 2010. PMID: 20368288 Free PMC article. - FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties.
Frey S, Görlich D. Frey S, et al. EMBO J. 2009 Sep 2;28(17):2554-67. doi: 10.1038/emboj.2009.199. Epub 2009 Aug 13. EMBO J. 2009. PMID: 19680227 Free PMC article. - A genomic glance at the components of the mRNA export machinery in Plasmodium falciparum.
Tuteja R, Mehta J. Tuteja R, et al. Commun Integr Biol. 2010 Jul;3(4):318-26. doi: 10.4161/cib.3.4.11886. Commun Integr Biol. 2010. PMID: 20798816 Free PMC article. - Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export.
Jani D, Valkov E, Stewart M. Jani D, et al. Nucleic Acids Res. 2014 Jun;42(10):6686-97. doi: 10.1093/nar/gku252. Epub 2014 Apr 4. Nucleic Acids Res. 2014. PMID: 24705649 Free PMC article. - OSBP-related protein 8 (ORP8) regulates plasma and liver tissue lipid levels and interacts with the nucleoporin Nup62.
Zhou T, Li S, Zhong W, Vihervaara T, Béaslas O, Perttilä J, Luo W, Jiang Y, Lehto M, Olkkonen VM, Yan D. Zhou T, et al. PLoS One. 2011;6(6):e21078. doi: 10.1371/journal.pone.0021078. Epub 2011 Jun 15. PLoS One. 2011. PMID: 21698267 Free PMC article.
References
- Allen N.P., Huang,L., Burlingame,A. and Rexach,M. (2001) Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem., 276, 29268–29274. - PubMed
- Bayliss R., Ribbeck,K., Akin,D., Kent,H.M., Feldherr,C.M., Görlich,D. and Stewart,M. (1999). Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol., 293, 579–593. - PubMed
- Bayliss R., Corbett,A.H. and Stewart,M. (2000a) The molecular mechanism of transport of macromolecules through nuclear pore complexes. Traffic, 1, 448–456. - PubMed
- Bayliss R., Littlewood,T.D. and Stewart,M. (2000b) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. - PubMed
- Bayliss R., Kent,H.M., Corbett,A.H. and Stewart,M. (2000c) Crystallization and initial X-ray diffraction characterization of complexes of FxFG nucleoporin repeats with nuclear transport factors. J. Struct. Biol., 131, 240–247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous