Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae - PubMed (original) (raw)
Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae
Hae-Sun Moon Park et al. Infect Immun. 2002 Jul.
Abstract
Expression of type IV pili (Tfp) correlates with the ability of Neisseria gonorrhoeae to colonize the human host, as well as with adherence to human epithelial tissue, twitching motility, competence for natural transformation, and autoagglutination. N. gonorrhoeae PilF (required for Tfp biogenesis) and PilT (required for twitching motility and transformation) share significant identities with members of a family of putative ATPases involved in membrane trafficking of macromolecules. An open reading frame downstream of the pilT locus encoding a 408-amino-acid protein with 33% identity with the gonococcal PilT protein and 45% identity with the PilU protein in Pseudomonas aeruginosa was characterized, and the corresponding gene was designated pilU. Unlike N. gonorrhoeae pilT mutants, pilU mutants express twitching motility and are competent for DNA transformation. However, loss-of-function mutations in pilU increased bacterial adherence to ME-180 human epithelial cells eightfold and disrupted in vitro Tfp-associated autoagglutination. Comparative alignment of N. gonorrhoeae PilU with other members of the TrbB-like family of traffic ATPases revealed a conserved carboxy-terminal domain unique to family members which are not essential for Tfp biogenesis but which specifically modify Tfp-associated phenotypes. Studies of the pilT-pilU locus by using Northern blotting, transcriptional fusions, and reverse transcription-PCR showed that the two genes encoding closely related proteins with dissimilar effects on Tfp phenotypes are transcribed from a single promoter.
Figures
FIG. 1.
Detection of PilU expressed in E. coli. Plasmid-encoded PilU and truncated PilU were expressed from the T7 promoter and analyzed by immunoblotting with affinity-purified anti-PilT rabbit serum. Whole-cell lysates were prepared from E. coli strain BL21 carrying the pT7-5 vector control, pT7U, or pT7U(Δ_Sal_I) grown in the absence (lanes 1, 3, and 5) or in the presence (lanes 2, 4, and 6) of isopropyl-β-
d
-thiogalactopyranoside (IPTG). Lanes 1 and 2, pT7-5; lanes 3 and 4, pT7U; lanes 5 and 6, pT7U(Δ_Sal_I).
FIG. 2.
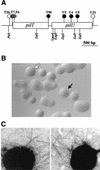
(A) Physical map of the gonococcal pilT-pilU locus. Transposon insertions are indicated by circles. The solid and striped circles indicate insertions resulting in PilU− and PilT− phenotypes, respectively, whereas the open circles indicate transposon insertions that had no effect on phenotypes. Restriction sites are also shown. (B) Colony morphologies of N. gonorrhoeae wild type (domed colony with a defined edge) (open arrow) and PilU− mutant (flat colony lacking a defined edge) (solid arrow). (C) Transmission electron micrographs of N. gonorrhoeae strains N400 (left panel) and GU2 (right panel). Other PilU− strains exhibited phenotypes identical to that exhibited by GU2. Bacteria were negatively stained with 0.5% ammonium molybdate. Magnification, ×39,000.
FIG. 3.
Quantitative analysis of Tfp and PilC in pilU mutants. The amounts of samples loaded were equalized based on the total protein concentration of whole cells from which Tfp were purified. (Top) Immunoblot of purified pili obtained by using rabbit antibodies specific for PilC. (Bottom) Coomassie blue-stained SDS-PAGE gel showing the relative amounts of PilE in purified pili.
FIG. 4.
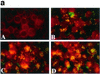
(a) Patterns of adherence of gonococcal strains to ME-180 cells. ME-180 cells were incubated for 1 h with bacteria, fixed, and stained with fluorescent conjugated monoclonal antibodies. (A) Nonpiliated strain GF2 (PilF−); (B) wild-type strain N400; (C) pilU mutant strain GU2; (D) pilU mutant strain GU5. (b) Increased adherence of PilU mutant strains to ME-180 cells. Adhesion assays were performed with ME-180 cells. The results are means of three experiments and are expressed as the ratios of the values for the mutants to the values for the wild-type control.
FIG. 4.
(a) Patterns of adherence of gonococcal strains to ME-180 cells. ME-180 cells were incubated for 1 h with bacteria, fixed, and stained with fluorescent conjugated monoclonal antibodies. (A) Nonpiliated strain GF2 (PilF−); (B) wild-type strain N400; (C) pilU mutant strain GU2; (D) pilU mutant strain GU5. (b) Increased adherence of PilU mutant strains to ME-180 cells. Adhesion assays were performed with ME-180 cells. The results are means of three experiments and are expressed as the ratios of the values for the mutants to the values for the wild-type control.
FIG. 5.
Amino acid sequence alignment of the carboxy-terminal segments of the GspE/TrbB family of proteins, determined by using Clustal X. Conserved residues are enclosed in boxes, and identical residues shaded. The alignment begins in the regions corresponding to conserved motif III as defined by Lessl and Lanka (top, underlined) (18), and numbering is based on the N. gonorrhoeae PilU sequence. The identical and similar residues unique to the PilT/PilU family members (dispensable for Tfp biogenesis but essential for associated phenotypes) begin after residue 250. This region contains the sequence motif GMQTXXXXLXXLXXXXXI (residues 313 to 333 of N. gonorrhoeae PilU). The following sequences are shown: NgPilT (N. gonorrhoeae, accession number AAB30824), PaPilT (P. aeruginosa, accession number P24559), MxPilT (M. xanthus, determined by Wu et al. [42]), SsPilT (Synechocystis sp., accession number BAA18564), SsPilT2 (Synechocystis sp., accession number BAA18443), AaPilT (Aquifex aeolicus, accession number AAC06903), NgPilU (N. gonorrhoeae, sequence determined in this study), PaPilU (P. aeruginosa, accession number S54702), EcBfpF (E. coli, accession number S70973), NgPilF (N. gonorrhoeae, accession number P37094), PaPilB (P. aeruginosa, accession number P22608), KpPulE (Klebsiella pneumoniae, accession number C34469), HiHofB (Haemophilus influenzae, accession number P44622), VcTcpT (V. cholerae, accession number P29480), and BsComGA (Bacillus subtilis, accession number B30338).
FIG. 6.
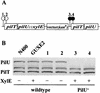
PilT and PilU expression in wild-type and mutant strains. (A) Physical map of the strains used. The solid and striped circles indicate transposon insertions resulting in PilU− and PilT− phenotypes, respectively. Names and sites of other mutations, as well as the restriction site created for construction of the pilTind allele (40), are also shown. (B) Immunoblot of whole-cell lysates from the wild type (lane 1), GTU2 (lane 2), GU21 (lane 3), GT2-17 (lane 4), GT101 (lane 5), GT102 (lane 6), GT50 (lane 7), and GT104 (pilTind) (lanes 7 and 8) with (lane +) or without (lane −) induction (200 μM isopropyl-β-
d
-thiogalactopyranoside). Immunoblots were probed with affinity-purified polyclonal anti-PilT serum.
FIG. 7.
Analysis of gene expression in the pilT-pilU locus with an xylE fusion construct. (A) Genomic configuration of strain GUXE2 created by integration of an xylE fusion plasmid (described in the text) into the genome of N400. The positions of transposon insertions in derivatives of strain GUXE2 which carry a duplication of the pilT gene and the flanking region are indicated by circles. The numbers above the circles correspond to lane numbers in panel B. (B) Immunoblot of whole-cell lysates from wild-type and fusion strains obtained by using affinity-purified polyclonal anti-PilT serum. The XylE activity was measured by the spray method as described in Materials and Methods, and the results were scored positive or negative. The phenotypes of each strain analyzed were noted.
FIG. 8.
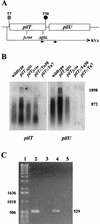
Transcription patterns within the _pilT_-pilU locus in wild-type and mutant strains. (A) Physical map of the locus in the strains analyzed. (B) Northern blot analyses of pilT and pilU expression. Total RNAs (10 μg) from wild-type and mutant strains were electrophoresed on a formaldehyde-agarose gel and blotted onto a nylon membrane. The transferred RNA was hybridized with a pilT probe, yielding the autoradiogram shown on the left. The membrane was then washed and rehybridized with the pilU probe, yielding the autoradiogram shown on the right. The positions of RNA size markers (in base pairs) are shown on the right. (C) Detection of an RNA species spanning pilT and pilU by RT-PCR. Lane 1, 1-kb DNA ladder; lane 2, positive control amplification of intergenic region with wild-type strain chromosomal DNA as the template and with the primers indicated by small arrows in panel A (the expected PCR product is 529 bp long); lane 3, negative control for RT-PCR, in which the conditions were identical to those used to obtain the product in lane 4, except that reverse transcriptase was not added to the reaction mixture; lane 4, RT-PCR with RNA from the wild-type strain; lane 5, RT-PCR with RNA from GT7.
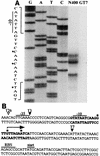
FIG. 9.
(A) Primer extension analysis of 5′ termini of pilT-pilU transcripts. RNA was isolated from strain N400 or GT7 after 5 h of culture in Gc liquid medium. Primer extension reaction mixtures were loaded alongside DNA sequencing reaction mixtures obtained by using wild-type genomic DNA templates with the extension primers. The letters above lanes G, A, T, and C (DNA sequencing reactions) indicate the dideoxynucleotides used to terminate the reactions. The nucleotide sequences surrounding the primer extension products have been expanded. The asterisks indicate nucleotides corresponding to the start points. (B) Nucleotide sequence of the upstream region of the pilT gene. Nucleotide sequences expanded in panel A are indicated by boldface type, and the transcriptional start sites mapped by primer extension analysis are indicated by asterisks. The arrow indicates the major pilT transcriptional start site. The consensus −10 and −35 hexamers, putative ribosome binding site for pilT (RBS), and translation start site are marked. The arrowheads show transposon insertion sites in mutant strains tested or discussed in the text. The solid arrowheads indicate a conferred pilT mutant phenotype, whereas the open arrowhead indicates that the transposon insertion had no effect.
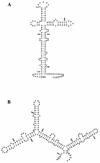
FIG. 10.
Predicted stem-loop structure of RNA derived from the _pilT_-pilU intergenic sequences generated by the FOLD program of the Wisconsin Genetics Computer Group package. The RNAs corresponding to the sense strand of the intergenic regions have optimal secondary structures with minimal free energies of −53.2 kcal/mol (length, 162 bp) for N. gonorrhoeae (A) and −64.1 kcal/mol (length, 171 bp) for P. aeruginosa (B). The numbering of nucleotides is as in GenBank accession number S72391 for panel A and M55524 for panel B. The asterisks indicate the stop codon of the pilT ORFs, and the dots indicate putative ribosome binding sites for pilU.
Similar articles
- Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa.
Whitchurch CB, Mattick JS. Whitchurch CB, et al. Mol Microbiol. 1994 Sep;13(6):1079-91. doi: 10.1111/j.1365-2958.1994.tb00499.x. Mol Microbiol. 1994. PMID: 7854122 - PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae.
Wolfgang M, Lauer P, Park HS, Brossay L, Hébert J, Koomey M. Wolfgang M, et al. Mol Microbiol. 1998 Jul;29(1):321-30. doi: 10.1046/j.1365-2958.1998.00935.x. Mol Microbiol. 1998. PMID: 9701824 - Type IV pili and cell motility.
Wall D, Kaiser D. Wall D, et al. Mol Microbiol. 1999 Apr;32(1):1-10. doi: 10.1046/j.1365-2958.1999.01339.x. Mol Microbiol. 1999. PMID: 10216854 Review. - Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer.
Koomey M. Koomey M. APMIS Suppl. 1998;84:56-61. doi: 10.1111/j.1600-0463.1998.tb05649.x. APMIS Suppl. 1998. PMID: 9850683 Review.
Cited by
- Type IV pilus retraction is required for Neisseria musculi colonization and persistence in a natural mouse model of infection.
Rhodes KA, Rendón MA, Ma MC, Agellon A, Johnson AC, So M. Rhodes KA, et al. mBio. 2024 Jan 16;15(1):e0279223. doi: 10.1128/mbio.02792-23. Epub 2023 Dec 12. mBio. 2024. PMID: 38084997 Free PMC article. - Type IV pili in Gram-positive bacteria.
Melville S, Craig L. Melville S, et al. Microbiol Mol Biol Rev. 2013 Sep;77(3):323-41. doi: 10.1128/MMBR.00063-12. Microbiol Mol Biol Rev. 2013. PMID: 24006467 Free PMC article. Review. - Type IV pilus assembly proficiency and dynamics influence pilin subunit phospho-form macro- and microheterogeneity in Neisseria gonorrhoeae.
Vik Å, Anonsen JH, Aas FE, Hegge FT, Roos N, Koomey M, Aspholm M. Vik Å, et al. PLoS One. 2014 May 5;9(5):e96419. doi: 10.1371/journal.pone.0096419. eCollection 2014. PLoS One. 2014. PMID: 24797914 Free PMC article. - The role of PilU in the surface behaviors of Pseudomonas aeruginosa.
Zhang J, Luo Y, Zong Y, Lu S, Shi Y, Jin F, Zhao K. Zhang J, et al. mLife. 2025 Feb 23;4(1):83-95. doi: 10.1002/mlf2.12165. eCollection 2025 Feb. mLife. 2025. PMID: 40026580 Free PMC article. - Canary in the Coal Mine: How Resistance Surveillance in Commensals Could Help Curb the Spread of AMR in Pathogenic Neisseria.
Goytia M, Wadsworth CB. Goytia M, et al. mBio. 2022 Oct 26;13(5):e0199122. doi: 10.1128/mbio.01991-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154280 Free PMC article. Review.
References
- Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941-951. - PubMed
- Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. - PubMed
- Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources