A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis - PubMed (original) (raw)
. 2002 Jun 27;417(6892):967-70.
doi: 10.1038/nature00769. Epub 2002 Jun 9.
Jennifer E G Gallagher, Patricia A Compagnone-Post, Brianna M Mitchell, Kara A Porwancher, Karen A Wehner, Steven Wormsley, Robert E Settlage, Jeffrey Shabanowitz, Yvonne Osheim, Ann L Beyer, Donald F Hunt, Susan J Baserga
Affiliations
- PMID: 12068309
- PMCID: PMC11487672
- DOI: 10.1038/nature00769
A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis
François Dragon et al. Nature. 2002.
Abstract
Although the U3 small nucleolar RNA (snoRNA), a member of the box C/D class of snoRNAs, was identified with the spliceosomal small nuclear RNAs (snRNAs) over 30 years ago, its function and its associated protein components have remained more elusive. The U3 snoRNA is ubiquitous in eukaryotes and is required for nucleolar processing of pre-18S ribosomal RNA in all organisms where it has been tested. Biochemical and genetic analyses suggest that U3 pre-rRNA base-pairing interactions mediate endonucleolytic pre-rRNA cleavages. Here we have purified a large ribonucleoprotein (RNP) complex from Saccharomyces cerevisiae that contains the U3 snoRNA and 28 proteins. Seventeen new proteins (Utp1 17) and Rrp5 were present, as were ten known components. The Utp proteins are nucleolar and specifically associated with the U3 snoRNA. Depletion of the Utp proteins impedes production of the 18S rRNA, indicating that they are part of the active pre-rRNA processing complex. On the basis of its large size (80S; calculated relative molecular mass of at least 2,200,000) and function, this complex may correspond to the terminal knobs present at the 5' ends of nascent pre-rRNAs. We have termed this large RNP the small subunit (SSU) processome.
Conflict of interest statement
Competing interests statement
The authors declare that they have no competing financial interests.
Figures
Figure 1
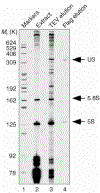
RNA composition of fractions from the purification of the U3 snoRNP. RNA was extracted from the starting material (extract, lane 2), from the first purification step that enriches for the box C/D snoRNAs (TEV elution, lane 3), and from the final eluate (Flag elution, lane 4). The RNA was directly labelled by 32p-labelled pCp and T4 RNA ligase, and analysed on an 8% denaturing polyacrylamide gel.
Figure 2
Function of the new components of the SSU processome. a, The Utp proteins are nucleolar. Anti-HA or anti-Myc antibodies were used to detect the tagged Utp proteins (red), whereas anti-Mpp10 antibodies were used to decorate the nucleolus (green). DAPI was used to stain the nucleus (blue). Utp proteins were triply tagged by HA or Myc. b, The Utp proteins and Rrp5 are complexed with Mpp10. Anti-HA immunoprecipitations performed on cell extracts were analysed for the presence of Mpp10 by western blotting with anti-Mpp10 antibodies. T, total (5% of the input for immunoprecipitation); IP, immunoprecipitate. c, The Utp proteins and Rrp5 are associated with U3 snoRNA. Anti-HA immunoprecipitations were performed on cell extracts and were washed at 150 mM (150) or 400 mM (400) NaCl. RNA was extracted and analysed for the presence of the U3 snoRNA by northern blotting. T, total (20% of the input); S, supernatant (20%). d, The Utp proteins are required for 18S rRNA biogenesis. Yeast strains conditionally expressing Utp proteins were grown in medium containing galactose/raffinose (0 h), but switched to glucose for protein depletion (24 or 48 h). RNA was extracted at the indicated time points and analysed for the presence of 25S and 18S rRNAs by northern blot.
Figure 3
The SSU processome sediments at 80S on sucrose gradients. Yeast extracts were analysed on 10–47% sucrose gradients. Anti-Mpp10, anti-Imp3, anti-Imp4 and anti-HA antibodies were used to detect the indicated proteins in each fraction by western blotting. Northern blotting was used to detect the U3 snoRNA. The migration of the 40S, 60S and 80S ribosomal subunits is specified. Utp16 and Nop1 proteins were triply tagged by HA.
Figure 4
The U3 snoRNA and Utp7 are required for terminal knob formation on nascent pre-rRNAs. Yeast strains conditionally expressing either the U3 snoRNA or Utp7 from a galactose promoter were used to make the chromatin spreads. Strains were undepleted (a) or depleted for U3 snoRNA (b) or Utp7 (c). For depletion, the strains were switched from growth in galactose to growth in glucose. Chromatin spreads were made before (0 h) and after (3 h) the switch to glucose and analysed by electron microscopy. Scale, the width of panel a is 0.85 μm.
Similar articles
- Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction.
Hoang T, Peng WT, Vanrobays E, Krogan N, Hiley S, Beyer AL, Osheim YN, Greenblatt J, Hughes TR, Lafontaine DL. Hoang T, et al. Mol Cell Biol. 2005 Jul;25(13):5523-34. doi: 10.1128/MCB.25.13.5523-5534.2005. Mol Cell Biol. 2005. PMID: 15964808 Free PMC article. - Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing.
Shah BN, Liu X, Correll CC. Shah BN, et al. RNA. 2013 Oct;19(10):1372-83. doi: 10.1261/rna.039511.113. Epub 2013 Aug 26. RNA. 2013. PMID: 23980203 Free PMC article. - Components of an interdependent unit within the SSU processome regulate and mediate its activity.
Wehner KA, Gallagher JE, Baserga SJ. Wehner KA, et al. Mol Cell Biol. 2002 Oct;22(20):7258-67. doi: 10.1128/MCB.22.20.7258-7267.2002. Mol Cell Biol. 2002. PMID: 12242301 Free PMC article. - Sno-capped: 5' ends of preribosomal RNAs are decorated with a U3 SnoRNP.
Culver GM. Culver GM. Chem Biol. 2002 Jul;9(7):777-9. doi: 10.1016/s1074-5521(02)00171-0. Chem Biol. 2002. PMID: 12144919 Review. - snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis.
Vos TJ, Kothe U. Vos TJ, et al. Cells. 2020 Sep 29;9(10):2195. doi: 10.3390/cells9102195. Cells. 2020. PMID: 33003357 Free PMC article. Review.
Cited by
- Noncoding RNAs in eukaryotic ribosome biogenesis and function.
Lafontaine DL. Lafontaine DL. Nat Struct Mol Biol. 2015 Jan;22(1):11-9. doi: 10.1038/nsmb.2939. Nat Struct Mol Biol. 2015. PMID: 25565028 Review. - Utp23p is required for dissociation of snR30 small nucleolar RNP from preribosomal particles.
Hoareau-Aveilla C, Fayet-Lebaron E, Jády BE, Henras AK, Kiss T. Hoareau-Aveilla C, et al. Nucleic Acids Res. 2012 Apr;40(8):3641-52. doi: 10.1093/nar/gkr1213. Epub 2011 Dec 17. Nucleic Acids Res. 2012. PMID: 22180534 Free PMC article. - atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana.
Weis BL, Palm D, Missbach S, Bohnsack MT, Schleiff E. Weis BL, et al. RNA. 2015 Mar;21(3):415-25. doi: 10.1261/rna.047563.114. Epub 2015 Jan 20. RNA. 2015. PMID: 25605960 Free PMC article. - The Contributions of the Ribosome Biogenesis Protein Utp5/WDR43 to Craniofacial Development.
Sondalle SB, Baserga SJ, Yelick PC. Sondalle SB, et al. J Dent Res. 2016 Oct;95(11):1214-20. doi: 10.1177/0022034516651077. Epub 2016 May 24. J Dent Res. 2016. PMID: 27221611 Free PMC article. Review. - Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding protein with a pronounced preference for U-rich sequences.
de Boer P, Vos HR, Faber AW, Vos JC, Raué HA. de Boer P, et al. RNA. 2006 Feb;12(2):263-71. doi: 10.1261/rna.2257606. RNA. 2006. PMID: 16428605 Free PMC article.
References
- Weinberg RA & Penman S Small molecular weight monodisperse nuclear RNA. J. Mol. Biol 38, 289–304 (1968). - PubMed
- Nakamura T, Prestayko AW & Busch H Studies on nucleolar 4 to 6S ribonucleic acid of Novikoff hepatoma cells. J. Biol. Chem 243, 1368–1375 (1968). - PubMed
- Venema J & Tollervey D Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet 33, 261–311 (1999). - PubMed
- Maxwell ES & Fournier MJ The small nucleolar RNAs. Ann. Rev. Biochem 64, 897–934 (1995). - PubMed
- Watkins NJ et al. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103, 457–466 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases