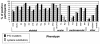Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes - PubMed (original) (raw)
Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes
Iris Schrijver et al. Am J Hum Genet. 2002 Aug.
Abstract
Marfan syndrome (MFS) and other type 1 fibrillinopathies result from mutations in the FBN1 gene, which encodes the connective-tissue microfibrillar protein fibrillin 1. Attempts at correlating genotype with phenotype have suggested considerable heterogeneity. To define the subtype of fibrillinopathy caused by premature termination codon (PTC) mutations, we integrate genotype information and mRNA expression levels with clinical and biochemical phenotypes. By screening the entire FBN1 gene for mutations, we identified 34 probands with PTC mutations. With the exception of two recurrent mutations, these nonsense and frameshift mutations are unique and span the entire FBN1 gene, from IVS2 to IVS63. Allele-specific reverse-transcriptase polymerase chain reaction analyses revealed differential allelic expression in all studied samples, with variable reduction of the mutant transcript. Fibrillin protein synthesis and deposition into the extracellular matrix were studied by pulse-chase analysis of cultured fibroblasts. In the majority of PTC samples, synthesis of normal-sized fibrillin protein was approximately 50% of control levels, but matrix deposition was disproportionately decreased. Probands and mutation-positive relatives were clinically evaluated by means of a standardized protocol. Only 71% (22/31) of probands and 58% (14/24) of the mutation-positive family members met current clinical diagnostic criteria for MFS. When compared with our previously reported study group of 44 individuals with FBN1 cysteine substitutions, the PTC group showed statistically significant differences in the frequency of individual signs, especially in the ocular manifestations. Whereas large-joint hypermobility was more common, lens dislocation and retinal detachment were distinctly less common in the PTC group. We conclude that PTC mutations have a major impact on the pathogenesis of type 1 fibrillinopathies and convey a distinct biochemical, clinical, and prognostic profile.
Figures
Figure 1
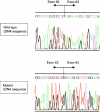
Effect of splice-site mutation 8051+1G→A on the FBN1 transcript. Direct sequencing of RT-PCR products from fibroblast of a normal control (top) and proband FB1477 with 8051+1G→A (bottom) reveals skipping of exon 63. This event leads to deletion of the two most C-terminal cbEGF-like domains and to a frameshift, resulting in premature termination of translation at amino acid position 2751 in exon 65 in the unique C-terminal region.
Figure 2
Detection of allele-specific transcripts. Top, Mutation 3778G→T abolishes a _Bpm_I site. Top left, Unlabeled RT-PCR of a region spanning nts 3506–3996 of the FBN1 cDNA sequence, digested with _Bpm_I. Digestion products from both the proband (P) and a normal control (C) are shown. Top right, 32P-labeled RT-PCR and digestion with _Bpm_I of proband cDNA. Both digested (D) and undigested (U) products are shown. Middle, Mutation 6185insA generates new _Mse_I site. Middle left, 32P-labeled RT-PCR and digestion with _Mse_I. Undigested (U) and digested (D) samples from proband are shown. Middle right, Genomic PCR of exon 50 from proband and digestion with _Mse_I. Bottom, Mutation 7240C→T abolishes _Bsl_I site. Bottom left, 32P labeled RT-PCR and digestion with _Bsl_I. Undigested (U) and digested (D) samples from proband are shown. Bottom right, 32P labeled genomic PCR of exon 58 from proband and digestion with _Bsl_I.
Figure 3
Comparison of transcript levels for FBN1 alleles distinguished by the C6236T _Rsa_I polymorphism in the 3′ UTR. All samples are from heterozygotes. RT-PCR products from fibroblast RNA samples were digested with _Rsa_I and electrophoresed on an SSCA gel under denaturing conditions. The band labeled “T” represents the uncut T allele, the two bands labeled “C” represent the cut C allele, and the band marked by an asterisk (*) is constant. Samples 3 and 12 are normal controls. Unequal transcript levels are evident in four samples with PTC mutations. Lane 1, FB 1058 (E1260X); lane 4, FB 1103 (L1161X); lane 6, FB773 (R1192X); and lane 11, FB997 (R1523X). Samples 1 and 11 reveal predominant transcripts with the C allele, whereas in samples 4 and 6 only the T allele is detectable. In contrast, the sample in lane 2 is from FB774, with a genomic deletion of exons 42 and 43 (Liu et al. 2001), and the one in lane 8 is from FB808, with the cysteine substitution C637R (Schrijver et al. 1999). Both of these samples, as well as the ones in lanes 5, 9, and 10, generated the normal pattern of fragments, indicating equal transcripts of both alleles. Lane 7 shows slight skewing towards the C allele. For samples 5, 7, 9, and 10, no FBN1 mutations have been identified as yet.
Figure 4
Reduced fibrillin 1 synthesis and deposition in a proband with a premature termination mutation in the FBN1 gene. Radioactively labeled fibrillin (arrows) is visualized after pulse-chase analysis of cultured skin fibroblasts from a normal control subject and FB1533 (5240insACACT, leading to a PTC in exon 46). After a 30-min pulse with 35S-cysteine, FB1533 demonstrates reduced synthesis of normal-sized fibrillin (34% of the normal control sample). Protein deposition is assessed after a 20-h “chase” period in which labeled fibrillin is secreted from the cells and accumulates in the extracellular matrix. Matrix deposition is reduced in FB1533. At 20% of the normal control level, it is disproportionately low, suggesting a dominant-negative mechanism. These results place FB1533 into protein group II (Aoyama et al. 1994).
Figure 5
Frequency of connective-tissue clinical features in study participants with PTC mutations, compared with those with cysteine substitutions (Schrijver et al. 1999). The percentage of all participants who are positive for the studied skeletal, ocular, cardiovascular, and other phenotypes in each of the two groups is demonstrated along the _Y_-axis. Individual manifestations are listed along the _X_-axis. Blackened columns represent PTC mutations, and shaded columns reflect cysteine substitutions. DCS = dolichostenomelia; PE/PC = pectus excavatum/pectus carinatum; SC = scoliosis; H/NP = high/narrow palate; DC = dental crowding; CTR = contractures; HSJ = hypermobile small joints; HLJ = hypermobile large joints; WS = wrist sign; TS = thumb sign; ARD = arachnodactyly; EL = ectopia lentis; RD = retinal detachment; AAA = ascending aortic aneurysm; AD = aortic dissection; ARR = aortic root replacement; MVP = mitral valve prolapse; MR = mitral regurgitation; and PMT = pneumothorax. Differences for HLJ, EL, and RD were statistically significant.
Similar articles
- Qualitative and quantitative analysis of FBN1 mRNA from 16 patients with Marfan Syndrome.
Tjeldhorn L, Amundsen SS, Barøy T, Rand-Hendriksen S, Geiran O, Frengen E, Paus B. Tjeldhorn L, et al. BMC Med Genet. 2015 Dec 18;16:113. doi: 10.1186/s12881-015-0260-4. BMC Med Genet. 2015. PMID: 26684006 Free PMC article. - Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes.
Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U. Schrijver I, et al. Am J Hum Genet. 1999 Oct;65(4):1007-20. doi: 10.1086/302582. Am J Hum Genet. 1999. PMID: 10486319 Free PMC article. - Identification of 29 novel and nine recurrent fibrillin-1 (FBN1) mutations and genotype-phenotype correlations in 76 patients with Marfan syndrome.
Rommel K, Karck M, Haverich A, von Kodolitsch Y, Rybczynski M, Müller G, Singh KK, Schmidtke J, Arslan-Kirchner M. Rommel K, et al. Hum Mutat. 2005 Dec;26(6):529-39. doi: 10.1002/humu.20239. Hum Mutat. 2005. PMID: 16220557 - Mutations of FBN1 and genotype-phenotype correlations in Marfan syndrome and related fibrillinopathies.
Robinson PN, Booms P, Katzke S, Ladewig M, Neumann L, Palz M, Pregla R, Tiecke F, Rosenberg T. Robinson PN, et al. Hum Mutat. 2002 Sep;20(3):153-61. doi: 10.1002/humu.10113. Hum Mutat. 2002. PMID: 12203987 Review. - Fibrillin-1 mutations in Marfan syndrome and other type-1 fibrillinopathies.
Hayward C, Brock DJ. Hayward C, et al. Hum Mutat. 1997;10(6):415-23. doi: 10.1002/(SICI)1098-1004(1997)10:6<415::AID-HUMU1>3.0.CO;2-C. Hum Mutat. 1997. PMID: 9401003 Review.
Cited by
- Identification of Genetic Variants Associated with Hereditary Thoracic Aortic Diseases (HTADs) Using Next Generation Sequencing (NGS) Technology and Genotype-Phenotype Correlations.
Butnariu LI, Russu G, Luca AC, Sandu C, Trandafir LM, Vasiliu I, Popa S, Ghiga G, Bălănescu L, Țarcă E. Butnariu LI, et al. Int J Mol Sci. 2024 Oct 17;25(20):11173. doi: 10.3390/ijms252011173. Int J Mol Sci. 2024. PMID: 39456956 Free PMC article. - Nonsense-mediated decay in genetic disease: friend or foe?
Miller JN, Pearce DA. Miller JN, et al. Mutat Res Rev Mutat Res. 2014 Oct-Dec;762:52-64. doi: 10.1016/j.mrrev.2014.05.001. Epub 2014 May 28. Mutat Res Rev Mutat Res. 2014. PMID: 25485595 Free PMC article. Review. - Qualitative and quantitative analysis of FBN1 mRNA from 16 patients with Marfan Syndrome.
Tjeldhorn L, Amundsen SS, Barøy T, Rand-Hendriksen S, Geiran O, Frengen E, Paus B. Tjeldhorn L, et al. BMC Med Genet. 2015 Dec 18;16:113. doi: 10.1186/s12881-015-0260-4. BMC Med Genet. 2015. PMID: 26684006 Free PMC article. - Pathogenic Mechanisms of Bicuspid Aortic Valve Aortopathy.
Yassine NM, Shahram JT, Body SC. Yassine NM, et al. Front Physiol. 2017 Sep 25;8:687. doi: 10.3389/fphys.2017.00687. eCollection 2017. Front Physiol. 2017. PMID: 28993736 Free PMC article. Review. - A new novel mutation in FBN1 causes autosomal dominant Marfan syndrome in a Chinese family.
Dong J, Bu J, Du W, Li Y, Jia Y, Li J, Meng X, Yuan M, Peng X, Zhou A, Wang L. Dong J, et al. Mol Vis. 2012;18:81-6. Epub 2012 Jan 13. Mol Vis. 2012. PMID: 22262941 Free PMC article.
References
Electronic-Database Information
- GeneTests, http://www.genetests.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MFS [MIM <154700>], FBN [MIM <113474>3], and APC [MIM <175100>])
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/
References
- Aoyama T, Francke U, Gasner C, Furthmayr H (1995) Fibrillin abnormalities and prognosis in Marfan syndrome and related disorders. Am J Med Genet 58:169–176 - PubMed
- Beroud C, Collod-Beroud G, Boileau C, Soussi T, Junien C (2000) UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum Mutat 15:86–94 - PubMed
- Biery NJ, Eldadah ZA, Moore CS, Stetten G, Spencer F, Dietz HC (1999) Revised genomic organization of FBN1 and significance for regulated gene expression. Genomics 56:70–77 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous