Overexpression of interleukin (IL)-7 leads to IL-15-independent generation of memory phenotype CD8+ T cells - PubMed (original) (raw)
Overexpression of interleukin (IL)-7 leads to IL-15-independent generation of memory phenotype CD8+ T cells
William C Kieper et al. J Exp Med. 2002.
Abstract
Transgenic (TG) mice expressing a high copy number of interleukin (IL)-7 cDNA under the control of the major histocomaptability complex (MHC) class II promoter display a 10-20-fold increase in total T cell numbers. Here, we show that the increase in T cell numbers in IL-7 TG mice is most apparent at the level of memory phenotype CD44hi CD122hi CD8+ cells. Based on studies with T cell receptor (TCR) TG mice crossed to IL-7 TG mice, increased levels of IL-7 may provide costimulation for TCR recognition of self-MHC ligands and thus cause naive CD8+ cells to proliferate and differentiate into memory phenotype cells. In addition, a marked increase in CD44hi CD122hi CD8+ cells was found in IL-7 TG IL-15(-) mice. Since these cell are rare in normal IL-15(-) mice, the dependency of memory phenotype CD8+ cells on IL-15 can be overcome by overexpression of IL-7.
Figures
Figure 1.
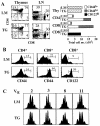
Characterization of T cells in young adult IL-7 TG mice. (A) T cell subset distribution and cell numbers in the thymus, LN, and spleen in IL-7 TG and LMs. Cells were triple-stained for CD4, CD8, CD44, or CD122 and analyzed by flow cytometry as described in Materials and Methods. Total numbers of thymocytes and indicated naive and memory phenotype CD4+ and CD8+ cells, combined from spleen and pooled LNs, IL-7 TG, and LMs are shown. Data are representative of >20 TG and LM mice. (B) Expression of CD44 and CD122 on gated LN CD4+ and CD8+ cells from IL-7 TG and LMs. (C) Randomly distributed CDR3 lengths of TCR-Vα chains expressed on CD8+ cells from IL-7 TG mice. CDR3 lengths on most Vα and all Vβ chains were determined on purified CD8+ cells from IL-7 TG and LM mice as described in Materials and Methods. Representative data for indicated Vα chains are shown; all analyzed chains were randomly distributed in both TG and LM cells.
Figure 2.
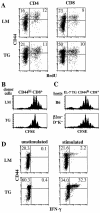
CD44hi CD8+ cells from IL-7 TG mice functionally resemble CD44hi CD8+ cells from wild-type mice. (A) Background turnover over of T cells IL-7 TG mice. Adult thymectomized IL-7 TG and LMs were given BrdU in the drinking water for 7 d and LN cells were triple stained for BrdU, CD44, and either CD4 or CD8 as described in Materials and Methods. (B) IL-7 TG CD44hi CD8+ cells undergo the same rate of homeostatic proliferation in T cell–depleted hosts as wild-type CD44hi CD8+ cells. CD44hi CD8+ cells from Ly 5.1+ IL-7 TG and LM mice purified by cell sorting were CFSE labeled and injected into irradiated (600 cGy) B6 mice. 7 d after transfer donor cells in host LN were analyzed for CFSE expression by staining for Ly5.1+ and CD8+. Shown are gated donor LMs and TG CD8+ cells. (C) Homeostatic proliferation of IL-7 TG CD44hi CD8+ cells is MHC independent. Sorted and CFSE-labeled CD44hi CD8+ cells from Ly 5.1+ IL-7 TG mice were injected into irradiated B6 and MHC class I− (β2m_–_D−K−) mice and analyzed 7 d later as in B. (D) IL-7 TG CD44hi CD8+ cells synthesize IFN-γ upon activation. Spleen cells from IL-7 TG and LMs were stimulated with anti-CD3 mAb in vitro for 7 h in the presence of Brefeldin A and stained for CD8, CD44, and intracellular IFN-γ as described in Materials and Methods. Shown are gated CD8+ cells. All data are representative of results from three experiments.
Figure 3.
Differential effect of IL-7 overproduction on OT-I and HY TCR TG CD8+ cells. OT-I and HY TCR TG mice were bred to express the IL-7 transgene; LN and spleen cells from 4-wk-old mice double TG mice were stained for CD8, Vα2, and CD44 for OT-I mice and CD8, T3.70, and CD44 for HY female mice. Shown on the left side for OT-I cells are the profiles of CD44 on gated CD8+ Vα2+ cells from normal OT-I TG mice (thin line) and IL-7 TG OT-I mice (thick line); CD44 levels on polyclonal CD8+ cells (broken line) from IL-7 TG mice are shown for comparison. For HY cells, shown are CD44 levels on gated T3.70+ cells from normal HY TG mice (thin line), IL-7 TG HY mice (thick line) and T3.70− cells from IL-7 TG HY mice (broken line). The total numbers (pooled from LN and spleen) of recovered CD44lo and CD44hi populations of CD8+ cells from the OT-I and HY mice are shown on the right side. Similar results were found when the cells were analyzed in terms of CD122 expression (not shown). Data are representative of results from four to six individuals for each type of mice.
Figure 4.
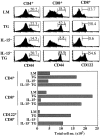
IL-15 is not required for generation of CD44hi CD8+ cells in IL-7 TG mice. LN cells from young adult IL-7 TG mice bred to an IL-15− background were stained for CD4, CD8, and CD44 or CD122 and analyzed. Shown are histograms of CD44 and CD122 on gated CD4+ and CD8+ cells as compared with the same cells from age-matched control LMs, IL-7 TG, and IL-15− mice. Total numbers of CD4+, CD8+, and CD122hi CD8+ cells from spleen and LNs from the mice are shown below. Data are representative of results from six to eight individuals for each type of mice.
Figure 5.
Role of IL-7 in supporting survival of CD44hi CD8+ cells in vitro and in vivo in IL-7 TG mice. (A) IL-7 can maintain survival of CD44hi CD8+ cells in vitro. Aliquots of purified IL-7 TG LN T cells were incubated with the indicated cytokines at (20 ng/ml) either alone or in the presence of anti–IL-7Rα mAb (A7R34), anti-CD122 mAb (TM-β1), or both mAbs at 50 μg/ml. The cells were harvested 5 d later and stained for CD4, CD8, and CD44, and the numbers of viable CD44hi CD8+ cells were calculated. (B) Blocking the IL-7R leads to depletion of CD44hi CD8+ cells in IL-7 TG mice. Groups of two to three LMs, IL-7 TG, and IL-15− IL-7 TG mice were injected three times with either 200 μg of anti–IL-7Rα mAb (A7R34) or rat IgG at 2-d intervals. 2 d after the last injection, LN and spleen cells were collected and stained for CD4, CD8, and CD44. The total numbers of CD44hi CD8+ cells recovered from spleens and pooled LN are shown. Another experiment showed similar results.
Comment in
- Multiple choices: regulation of memory CD8 T cell generation and homeostasis by interleukin (IL)-7 and IL-15.
Prlic M, Lefrancois L, Jameson SC. Prlic M, et al. J Exp Med. 2002 Jun 17;195(12):F49-52. doi: 10.1084/jem.20020767. J Exp Med. 2002. PMID: 12070294 Free PMC article. No abstract available.
Similar articles
- Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells.
Tan JT, Ernst B, Kieper WC, LeRoy E, Sprent J, Surh CD. Tan JT, et al. J Exp Med. 2002 Jun 17;195(12):1523-32. doi: 10.1084/jem.20020066. J Exp Med. 2002. PMID: 12070280 Free PMC article. - IL-6, in synergy with IL-7 or IL-15, stimulates TCR-independent proliferation and functional differentiation of CD8+ T lymphocytes.
Gagnon J, Ramanathan S, Leblanc C, Cloutier A, McDonald PP, Ilangumaran S. Gagnon J, et al. J Immunol. 2008 Jun 15;180(12):7958-68. doi: 10.4049/jimmunol.180.12.7958. J Immunol. 2008. PMID: 18523259 - Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells.
Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, Butz EA. Goldrath AW, et al. J Exp Med. 2002 Jun 17;195(12):1515-22. doi: 10.1084/jem.20020033. J Exp Med. 2002. PMID: 12070279 Free PMC article. - IL-7: maintaining T-cell memory and achieving homeostasis.
Bradley LM, Haynes L, Swain SL. Bradley LM, et al. Trends Immunol. 2005 Mar;26(3):172-6. doi: 10.1016/j.it.2005.01.004. Trends Immunol. 2005. PMID: 15745860 Review. - Homeostasis of Naive and Memory T Lymphocytes.
Kawabe T, Yi J, Sprent J. Kawabe T, et al. Cold Spring Harb Perspect Biol. 2021 Sep 1;13(9):a037879. doi: 10.1101/cshperspect.a037879. Cold Spring Harb Perspect Biol. 2021. PMID: 33753403 Free PMC article. Review.
Cited by
- Memory T cells in transplantation - progress and challenges.
Li XC, Kloc M, Ghobrial RM. Li XC, et al. Curr Opin Organ Transplant. 2013 Aug;18(4):387-92. doi: 10.1097/MOT.0b013e3283626130. Curr Opin Organ Transplant. 2013. PMID: 23838642 Free PMC article. Review. - Cytokine-Mediated Regulation of CD8 T-Cell Responses During Acute and Chronic Viral Infection.
Hashimoto M, Im SJ, Araki K, Ahmed R. Hashimoto M, et al. Cold Spring Harb Perspect Biol. 2019 Jan 2;11(1):a028464. doi: 10.1101/cshperspect.a028464. Cold Spring Harb Perspect Biol. 2019. PMID: 29101105 Free PMC article. Review. - Enhanced expression of cell cycle regulatory genes in virus-specific memory CD8+ T cells.
Latner DR, Kaech SM, Ahmed R. Latner DR, et al. J Virol. 2004 Oct;78(20):10953-9. doi: 10.1128/JVI.78.20.10953-10959.2004. J Virol. 2004. PMID: 15452215 Free PMC article. - IL-7 promotes the transition of CD4 effectors to persistent memory cells.
Li J, Huston G, Swain SL. Li J, et al. J Exp Med. 2003 Dec 15;198(12):1807-15. doi: 10.1084/jem.20030725. J Exp Med. 2003. PMID: 14676295 Free PMC article. - Low IL7R Expression at Diagnosis Predicted Relapse in Adult Acute Myeloid Leukemia Patients With t(8;21).
Xu N, Sun K, Wang YZ, Chen WM, Wang J, Li LD, Wang X, Hao Y, Chang Y, Liu YR, Huang XJ, Qin YZ. Xu N, et al. Front Immunol. 2022 Jul 7;13:909104. doi: 10.3389/fimmu.2022.909104. eCollection 2022. Front Immunol. 2022. PMID: 35874754 Free PMC article.
References
- Goldrath, A.W., and M.J. Bevan. 1999. Selecting and maintaining a diverse T-cell repertoire. Nature. 402:255–262. - PubMed
- Ernst, B., D.-S. Lee, J.M. Chang, J. Sprent, and C.D. Surh. 1999. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 11:173–181. - PubMed
- Schluns, K.S., W.C. Kieper, S.C. Jameson, and L. Lefrancois. 2000. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1:426–432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA38355/CA/NCI NIH HHS/United States
- AI21487/AI/NIAID NIH HHS/United States
- AI46710/AI/NIAID NIH HHS/United States
- HL07196/HL/NHLBI NIH HHS/United States
- R01 AI046710/AI/NIAID NIH HHS/United States
- R37 CA038355/CA/NCI NIH HHS/United States
- AG20186/AG/NIA NIH HHS/United States
- R01 AI045809/AI/NIAID NIH HHS/United States
- T32 AI007244/AI/NIAID NIH HHS/United States
- AI07244/AI/NIAID NIH HHS/United States
- R01 AG020186/AG/NIA NIH HHS/United States
- AI45809/AI/NIAID NIH HHS/United States
- AI41079/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous