Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments - PubMed (original) (raw)
Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments
Matthew T Webster et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Most studies of microsatellite evolution utilize long, highly mutable loci, which are unrepresentative of the majority of simple repeats in the human genome. Here we use an unbiased sample of 2,467 microsatellite loci derived from alignments of 5.1 Mb of genomic sequence from human and chimpanzee to investigate the mutation process of tandemly repetitive DNA. The results indicate that the process of microsatellite evolution is highly heterogeneous, exhibiting differences between loci of different lengths and motif sizes and between species. We find a highly significant tendency for human dinucleotide repeats to be longer than their orthologues in chimpanzees, whereas the opposite trend is observed in mononucleotide repeat arrays. Furthermore, the rate of divergence between orthologues is significantly higher at longer loci, which also show significantly greater mutability per repeat number. These observations have important consequences for understanding the molecular mechanisms of microsatellite mutation and for the development of improved measures of genetic distance.
Figures
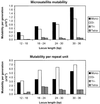
Figure 1
Effect of allele length on mutability per generation measured per locus (Upper) and per repeat unit (Lower) for microsatellites of repeat motif sizes 1–4 bp. Loci with allele lengths on the boundary between classes were placed in the higher category.
Figure 2
Results of simulations of microsatellite evolution in two species with constant mutation rates independent of allele length. Alleles were sampled from the final distribution on the basis of length in one species and a correction for unobserved reverse mutations was performed. Average mutability is plotted relative to its expected value given the simulated mutation rate. Both high (0.05 per generation) and low (0.0005 per generation) mutation rates were simulated, leading to expected average squared differences of 1 and 100, respectively.
Similar articles
- Divergent microsatellite evolution in the human and chimpanzee lineages.
Gáspári Z, Ortutay C, Tóth G. Gáspári Z, et al. FEBS Lett. 2007 May 29;581(13):2523-6. doi: 10.1016/j.febslet.2007.04.073. Epub 2007 May 4. FEBS Lett. 2007. PMID: 17498704 - The genome-wide determinants of human and chimpanzee microsatellite evolution.
Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. Kelkar YD, et al. Genome Res. 2008 Jan;18(1):30-8. doi: 10.1101/gr.7113408. Epub 2007 Nov 21. Genome Res. 2008. PMID: 18032720 Free PMC article. - Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size.
Garza JC, Slatkin M, Freimer NB. Garza JC, et al. Mol Biol Evol. 1995 Jul;12(4):594-603. doi: 10.1093/oxfordjournals.molbev.a040239. Mol Biol Evol. 1995. PMID: 7659015 - [Comparative studies on human and chimpanzee genomes].
Yoko K, Atsushi T, Hideki N, Asao F. Yoko K, et al. Tanpakushitsu Kakusan Koso. 2005 Dec;50(16 Suppl):2072-7. Tanpakushitsu Kakusan Koso. 2005. PMID: 16411432 Review. Japanese. No abstract available. - [Chimpanzee genome sequencing and comparative analysis with the human genome].
Watanabe H, Hattori M. Watanabe H, et al. Tanpakushitsu Kakusan Koso. 2006 Feb;51(2):178-87. Tanpakushitsu Kakusan Koso. 2006. PMID: 16457209 Review. Japanese. No abstract available.
Cited by
- A versatile microsatellite instability reporter system in human cells.
Koole W, Schäfer HS, Agami R, van Haaften G, Tijsterman M. Koole W, et al. Nucleic Acids Res. 2013 Sep;41(16):e158. doi: 10.1093/nar/gkt615. Epub 2013 Jul 16. Nucleic Acids Res. 2013. PMID: 23861444 Free PMC article. - Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome.
Liu G; NISC Comparative Sequencing Program; Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE. Liu G, et al. Genome Res. 2003 Mar;13(3):358-68. doi: 10.1101/gr.923303. Genome Res. 2003. PMID: 12618366 Free PMC article. - Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels.
Britten RJ. Britten RJ. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13633-5. doi: 10.1073/pnas.172510699. Epub 2002 Oct 4. Proc Natl Acad Sci U S A. 2002. PMID: 12368483 Free PMC article. - Comparative genetics of functional trinucleotide tandem repeats in humans and apes.
Andrés AM, Soldevila M, Lao O, Volpini V, Saitou N, Jacobs HT, Hayasaka I, Calafell F, Bertranpetit J. Andrés AM, et al. J Mol Evol. 2004 Sep;59(3):329-39. doi: 10.1007/s00239-004-2628-5. J Mol Evol. 2004. PMID: 15553088 - Tandem repeat variation in human and great ape populations and its impact on gene expression divergence.
Bilgin Sonay T, Carvalho T, Robinson MD, Greminger MP, Krützen M, Comas D, Highnam G, Mittelman D, Sharp A, Marques-Bonet T, Wagner A. Bilgin Sonay T, et al. Genome Res. 2015 Nov;25(11):1591-9. doi: 10.1101/gr.190868.115. Epub 2015 Aug 19. Genome Res. 2015. PMID: 26290536 Free PMC article.
References
- Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. - PubMed
- Goldstein D B, Schlötterer C, editors. Microsatellites: Evolution and Applications. Oxford: Oxford Univ. Press; 1999.
- Stallings R L, Ford A F, Nelson D, Torney D C, Hildebrand C E, Moyzis R K. Genomics. 1991;10:807–815. - PubMed
- Neff B D, Gross M R. Evolution (Lawrence, Kans) 2001;55:1717–1733. - PubMed
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature (London) 1996;380:152–154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous