A comprehensive analysis of recently integrated human Ta L1 elements - PubMed (original) (raw)
doi: 10.1086/341718. Epub 2002 Jun 17.
Bethaney J Vincent, Hunt Udall, W Scott Watkins, Tammy A Morrish, Gail E Kilroy, Gary D Swergold, Jurgen Henke, Lotte Henke, John V Moran, Lynn B Jorde, Mark A Batzer
Affiliations
- PMID: 12070800
- PMCID: PMC379164
- DOI: 10.1086/341718
A comprehensive analysis of recently integrated human Ta L1 elements
Jeremy S Myers et al. Am J Hum Genet. 2002 Aug.
Abstract
The Ta (transcribed, subset a) subfamily of L1 LINEs (long interspersed elements) is characterized by a 3-bp ACA sequence in the 3' untranslated region and contains approximately 520 members in the human genome. Here, we have extracted 468 Ta L1Hs (L1 human specific) elements from the draft human genomic sequence and screened individual elements using polymerase-chain-reaction (PCR) assays to determine their phylogenetic origin and levels of human genomic diversity. One hundred twenty-four of the elements amenable to complete sequence analysis were full length ( approximately 6 kb) and have apparently escaped any 5' truncation. Forty-four of these full-length elements have two intact open reading frames and may be capable of retrotransposition. Sequence analysis of the Ta L1 elements showed a low level of nucleotide divergence with an estimated age of 1.99 million years, suggesting that expansion of the L1 Ta subfamily occurred after the divergence of humans and African apes. A total of 262 Ta L1 elements were screened with PCR-based assays to determine their phylogenetic origin and the level of human genomic variation associated with each element. All of the Ta L1 elements analyzed by PCR were absent from the orthologous positions in nonhuman primate genomes, except for a single element (L1HS72) that was also present in the common (Pan troglodytes) and pygmy (P. paniscus) chimpanzee genomes. Sequence analysis revealed that this single exception is the product of a gene conversion event involving an older preexisting L1 element. One hundred fifteen (45%) of the Ta L1 elements were polymorphic with respect to insertion presence or absence and will serve as identical-by-descent markers for the study of human evolution.
Figures
Figure 1
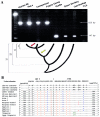
Human diversity associated with a truncated Ta L1Hs element, as shown by an agarose gel chromatograph of the PCR products from a survey of the human genomic variation associated with L1HS7. Amplification of the pre-integration site of this locus generates a 130-bp PCR product; amplification of a filled site generates a 326-bp product (by use of flanking unique sequence primers). In this survey of human genomic variation, 20 individuals from each of four diverse populations were assayed for the presence or absence of the L1 element, with only the African American samples shown here; the control samples (gray lines) were TLE buffer (i.e., 10 mM Tris-HCl:0.1 mM EDTA), common chimpanzee, gorilla, and owl monkey DNA templates. Most of the individuals surveyed were homozygous for the presence of the L1 element; in addition, this particular L1 element was absent from the genomes of nonhuman primates.
Figure 2
Human diversity associated with a long L1Hs Ta insertion polymorphism, as shown by an agarose gel chromatograph of the PCR products from a survey of the human genomic variation associated with L1HS364. Because of the size (∼6,000 bp) of this L1 element, two separate PCRs are performed to genotype individual samples. In the first reaction, flanking unique sequence primers were used to genotype the empty alleles (A); amplification of empty alleles from this locus generates a 97-bp PCR product. In the second reaction, a Ta subfamily–specific internal primer termed “ACA” and the 3′ flanking unique sequence primer were used to genotype filled sites (B); the amplification of filled sites generates a 170-bp product. In this survey of human genomic variation, 20 individuals from each of four diverse populations were assayed for the presence or absence of the L1 element, with only the Egyptian samples shown here; the control samples (black lines) were TLE buffer, common chimpanzee, gorilla, and owl monkey DNA templates. This particular L1 insertion polymorphism is a high-frequency insertion polymorphism, and most of the individuals surveyed have L1 filled chromosomes.
Figure 3
L1HS72 gene conversion. A, Agarose gel chromatograph of the PCR products derived from the amplification of L1HS72 in a series of human and nonhuman primate genomes, with a schematic of the primate evolutionary tree over the past 35 million years shown below. The yellow notched arrow represents the approximate time period when the L1HS72 element first integrated, and the red notched arrow represents the approximate time period of the gene conversion event of the preexisting L1 element. The fragment-length marker is a 123-bp ladder. B, Sequence alignment generated by sequencing the L1HS72 amplicons from nine diverse humans. Sequences are compared relative to L1Hs Ta consensus sequence and the L1HS72 sequence obtained from GenBank with only the diagnostic bases shown and positions reported relative to L1 retrotransposable element–1 (Dombroski et al. 1991). The G and C at positions 5536 and 5539 are indicative of the Ta-0 subset, whereas the Ta-1 subset has T and G at these nucleotides (Boissinot et al. 2000). The G at position 6015 (in addition to the ACA at positions 5930–5932) is diagnostic for the L1Hs Ta subfamily (Ovchinnikov et al. 2001). The target-site duplication sequence (TSD) is shown in brackets. The mosaic elements seen in the human samples are believed to be the result of at least one gene conversion, some time after the divergence of humans from the great apes (approximately five million years ago), of a preexisting L1 element with a younger L1Hs element. In the representation of nucleotides, different colors are used to denote conserved sequences and sequence variations between samples: green denotes bases unique to the common and pygmy chimpanzee genomes; blue denotes nucleotides unique to the human samples; orange denotes shared bases conserved between the common chimpanzee, pygmy chimpanzee, and human samples; and red denotes SNPs, within L1HS72, in the human population.
Figure 4
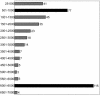
Ta L1 element size classes (in bp), showing the size distribution of Ta L1Hs elements. Elements are grouped in 500-bp intervals ranging from <500 bp to 7,000 bp long. The two most common size intervals are shown in black.
Figure 5
L1HS169-mediated transduction, showing an L1Hs transduction event. L1HS169 marked by clear target-site duplications is the putative source gene for L1HS28. The L1HS28 insertion contains 3′ flanking sequences identical to that of L1HS169 and unique target-site duplications flanking this entire sequence—suggesting that L1HS28 was created from a read-through transcript of L1HS169 that, to give rise to L1HS28, integrated into a new location on chromosome X. In addition, a second transduction event—L1HS547, from chromosome 18—is also flanked by unique target-site duplications and was also derived from L1HS169.
Similar articles
- LINE-1 preTa elements in the human genome.
Salem AH, Myers JS, Otieno AC, Watkins WS, Jorde LB, Batzer MA. Salem AH, et al. J Mol Biol. 2003 Feb 28;326(4):1127-46. doi: 10.1016/s0022-2836(03)00032-9. J Mol Biol. 2003. PMID: 12589758 - Following the LINEs: an analysis of primate genomic variation at human-specific LINE-1 insertion sites.
Vincent BJ, Myers JS, Ho HJ, Kilroy GE, Walker JA, Watkins WS, Jorde LB, Batzer MA. Vincent BJ, et al. Mol Biol Evol. 2003 Aug;20(8):1338-48. doi: 10.1093/molbev/msg146. Epub 2003 May 30. Mol Biol Evol. 2003. PMID: 12777507 - Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition.
Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Sheen FM, et al. Genome Res. 2000 Oct;10(10):1496-508. doi: 10.1101/gr.149400. Genome Res. 2000. PMID: 11042149 Free PMC article. - Alu repeats and human genomic diversity.
Batzer MA, Deininger PL. Batzer MA, et al. Nat Rev Genet. 2002 May;3(5):370-9. doi: 10.1038/nrg798. Nat Rev Genet. 2002. PMID: 11988762 Review. - A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease.
Chen JM, Stenson PD, Cooper DN, Férec C. Chen JM, et al. Hum Genet. 2005 Sep;117(5):411-27. doi: 10.1007/s00439-005-1321-0. Epub 2005 Jun 28. Hum Genet. 2005. PMID: 15983781 Review.
Cited by
- Altering Genomic Integrity: Heavy Metal Exposure Promotes Transposable Element-Mediated Damage.
Morales ME, Servant G, Ade C, Roy-Engel AM. Morales ME, et al. Biol Trace Elem Res. 2015 Jul;166(1):24-33. doi: 10.1007/s12011-015-0298-3. Epub 2015 Mar 14. Biol Trace Elem Res. 2015. PMID: 25774044 Free PMC article. Review. - Analysis of the human Alu Ye lineage.
Salem AH, Ray DA, Hedges DJ, Jurka J, Batzer MA. Salem AH, et al. BMC Evol Biol. 2005 Feb 22;5:18. doi: 10.1186/1471-2148-5-18. BMC Evol Biol. 2005. PMID: 15725352 Free PMC article. - Analysis of 5' junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5'-end attachment requiring microhomology-mediated end-joining.
Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Löwer J, Schumann GG. Zingler N, et al. Genome Res. 2005 Jun;15(6):780-9. doi: 10.1101/gr.3421505. Genome Res. 2005. PMID: 15930490 Free PMC article. - Why are young and old repetitive elements distributed differently in the human genome?
Belle EM, Webster MT, Eyre-Walker A. Belle EM, et al. J Mol Evol. 2005 Mar;60(3):290-6. doi: 10.1007/s00239-004-0020-0. J Mol Evol. 2005. PMID: 15871040 - L1 antisense promoter drives tissue-specific transcription of human genes.
Mätlik K, Redik K, Speek M. Mätlik K, et al. J Biomed Biotechnol. 2006;2006(1):71753. doi: 10.1155/JBB/2006/71753. J Biomed Biotechnol. 2006. PMID: 16877819 Free PMC article.
References
Electronic-Database Information
- Batzer Lab, http://batzerlab.lsu.edu/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the DNA sequences from the common and pygmy chimpanzee orthologs of L1HS72 [accession numbers AF489459 and AF489460]; diverse DNA sequences from L1HS72 [accession numbers AF489450–AF489458]; and Ta L1 element pre-integration site sequences, namely, L1HS45 [accession numbers AF461364 and AF461365], L1HS172 [accession numbers AF461368 and AF461369], L1HS178 [accession numbers AF461370 and AF461371], L1HS284 [accession numbers AF461372 and AF461373], L1HS372 [accession numbers AF461374 and AF461375], L1HS416 [accession numbers AF461376 and AF461377], L1HS442 [accession numbers AF461378 and AF461379], L1HS443 [accession numbers AF461386 and AF461387], L1HS513 [accession numbers AF461380–AF461382], and L1HS558 [accession number AF461383])
- Genetic Information Research Institute Censor Server, http://www.girinst.org/Censor_Server-Data_Entry_Forms.html
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 - PubMed
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA (1995) Alu repeats: a source for the genesis of primate microsatellites. Genomics 29:136–144 - PubMed
- Ausabel FM, Brent R, Kingston ME, Moore DD, Seidman JG (1987) Current protocols in molecular biology. John Wiley & Sons, New York
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM 59290/GM/NIGMS NIH HHS/United States
- R01 GM060518/GM/NIGMS NIH HHS/United States
- R21 CA 87356-02/CA/NCI NIH HHS/United States
- R01 GM 60518/GM/NIGMS NIH HHS/United States
- R01 GM059290/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials