An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells - PubMed (original) (raw)
An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells
Lisa L Hall et al. Proc Natl Acad Sci U S A. 2002.
Abstract
It has been believed that XIST RNA requires a discrete window in early development to initiate the series of chromatin-remodeling events that form the heterochromatic inactive X chromosome. Here we investigate four adult male HT-1080 fibrosarcoma cell lines expressing ectopic human XIST and demonstrate that these postdifferentiation cells can undergo chromosomal inactivation outside of any normal developmental context. All four clonal lines inactivated the transgene-containing autosome to varying degrees and with variable stability. One clone in particular consistently localized the ectopic XIST RNA to a discrete chromosome territory that exhibited striking hallmarks of inactivation, including long-range transcriptional inactivation. Results suggest that some postdifferentiation cell lines are capable of de novo chromosomal inactivation; however, long-term retention of autosomal inactivation was less common, which suggests that autosomal inactivation may confer a selective disadvantage. These results have fundamental significance for understanding genomic programming in early development.
Figures
Figure 1
XIST construct. (Top) Diagram represents genomic DNA spanning the human XIST gene. Cosmids P1644 and F235 contain the entire XIST gene (30 kb) and a β-geo selection marker. (Middle) P1644 includes ≈10 kb of 5′ flanking sequence, and (Bottom) F235 includes ≈10 kb of 3′ DNA. The F1–2 and F2–6 clones have the F235 construct, and the P1 clone has the P1644 construct. F1–2 has the β-geo marker in one orientation (F1) and the F2–6 line has the opposite orientation (F2).
Figure 2
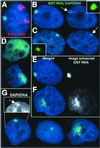
XIST RNA localization and Barr body formation. (A) The F2–6 TC chromosome is not the X chromosome. Ectopic XIST RNA (green), X chromosome paint (red). (B) F2–6 XIST RNA (green) and condensed DNA of a Barr body (arrow) (in 98% of expressing cells). (C) XIST RNA (green) and Xi Barr body (arrow) in normal female WI-38 cells. (D) Some F1–2 cells (5%) show large XIST RNA (green) drift. Most (70%) show diffuse cloud. (E and F) The XIST RNA (green) signal is enhanced equally for F1–2 and F2–6 cells (black and white images). XIST RNA drift can be seen in the more focal F1–2 cells (F). Equally enhanced F2–6 signal (E) shows little drift. (G) The P1 clone shows variable XIST localization: 74% are more localized, 20% have small spots, and 6% have large clouds. The more localized signal also shows some DNA condensation (arrow, Inset). (Magnification: ×2,000.)
Figure 3
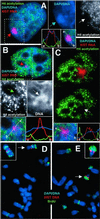
Acetylation and late-replication analysis. (A_–_C) XIST RNA probe (red) and α-acetylated H4 antibody (green). (A) F2–6 cells reveal a hole (white arrow) in the acetylation over the XIST RNA (red arrow) and Barr body (blue arrow). Some diminished signal is seen in both P1 (B; green arrow) and F1–2 (C; arrow). Fluorescence intensity plots across the XIST signal (white line in merged Insets) illustrate the acetylation holes. Absence of acetylation in these cell lines sometimes corresponds to DNA condensation (B; white arrow). (D) F2–6 metaphase spread show XIST (red) and BrdUrd (green) (5 h BrdUrd sample). (Inset) The TC chromosome from a slightly earlier-replicating spread exhibiting a third late-replicating band on the p arm. The single copy endogenous XIST gene does not appear in these images because the signal intensity of the ectopic XIST is so much higher. (E) Some F1–2 cells (40%) show a late-replicating chromosome, (arrow). (Inset) An example of the same chromosome from another spread. (Magnifications: ×2,000, A–C; ×3,500, D and E.)
Figure 4
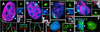
Global transcription analysis. (A_–_D) Hybridization of Cot-1 DNA probe to predominantly intron containing heterogeneous nuclear RNA (red) and XIST RNA (green). Both the WI-38 normal female (A) and the F2–6 (B) lines show a hole (red arrows) in the Cot-1 signal coincident with the XIST RNA (green signal), and Barr body (arrow, blue insets). Fluorescence intensity plots for each signal across the area of the XIST localization (white line in merged image) illustrate the transcription holes coincident with the XIST signal and Barr bodies. The measurement line in the F2–6 cell (B) was curved to avoid the large Cot1 signal. Both F1–2 (C) and P1 (D) cells usually show diminished transcription (red arrows, Insets) under the brightest XIST signal. DNA condensation is sometimes associated with the transcription hole (C; arrow, blue insets). (E and F) F2–6 cells. BrUTP incorporation (green) into newly transcribed RNA shows a hole (white arrows) in the transcription signal coincident with the Barr body and the ectopic XIST gene (E; red arrow). (Magnifications: ×2,000, A–E; ×1,500, F.)
Figure 5
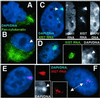
State of differentiation and selection against inactivation. (A) F1–2 and (B) F2–6 cells contain cytoplasmic cytokeratins (green). A TC Barr body can be seen in the F2–6 cell (B) (arrow). (C) ≈1% of F2–6 cells exhibit two sites of XIST integration. Insets reveal only the top site (arrow) has a transcription hole, good RNA localization, and a clear Barr body. (D) Earlier passage F1–2 cells showed better localization of XIST RNA (green) and DNA condensation (arrow, DNA Inset). (E and F) L1.10.1 cells with localized XIST RNA (red) (E) and Barr bodies (arrow, DNA Inset). Localization was lost during propagation (F Inset) in all of the cells. (Magnifications: ×1,500, A, B, and D; ×2,000, C and F.)
Similar articles
- Unbalanced X;autosome translocations provide evidence for sequence specificity in the association of XIST RNA with chromatin.
Hall LL, Clemson CM, Byron M, Wydner K, Lawrence JB. Hall LL, et al. Hum Mol Genet. 2002 Dec 1;11(25):3157-65. doi: 10.1093/hmg/11.25.3157. Hum Mol Genet. 2002. PMID: 12444100 - Xist has properties of the X-chromosome inactivation centre.
Herzing LB, Romer JT, Horn JM, Ashworth A. Herzing LB, et al. Nature. 1997 Mar 20;386(6622):272-5. doi: 10.1038/386272a0. Nature. 1997. PMID: 9069284 - Xist RNA and the mechanism of X chromosome inactivation.
Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Plath K, et al. Annu Rev Genet. 2002;36:233-78. doi: 10.1146/annurev.genet.36.042902.092433. Epub 2002 Jun 11. Annu Rev Genet. 2002. PMID: 12429693 Review. - Heterochromatin.
Hennig W. Hennig W. Chromosoma. 1999 Apr;108(1):1-9. doi: 10.1007/s004120050346. Chromosoma. 1999. PMID: 10199951 Review.
Cited by
- Localization of RNAs in the nucleus: _cis_- and _trans_- regulation.
Tong C, Yin Y. Tong C, et al. RNA Biol. 2021 Dec;18(12):2073-2086. doi: 10.1080/15476286.2021.1894025. Epub 2021 Mar 8. RNA Biol. 2021. PMID: 33682620 Free PMC article. Review. - Two-step imprinted X inactivation: repeat versus genic silencing in the mouse.
Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Namekawa SH, et al. Mol Cell Biol. 2010 Jul;30(13):3187-205. doi: 10.1128/MCB.00227-10. Epub 2010 Apr 19. Mol Cell Biol. 2010. PMID: 20404085 Free PMC article. - Xist spatially amplifies SHARP/SPEN recruitment to balance chromosome-wide silencing and specificity to the X chromosome.
Jachowicz JW, Strehle M, Banerjee AK, Blanco MR, Thai J, Guttman M. Jachowicz JW, et al. Nat Struct Mol Biol. 2022 Mar;29(3):239-249. doi: 10.1038/s41594-022-00739-1. Epub 2022 Mar 17. Nat Struct Mol Biol. 2022. PMID: 35301492 Free PMC article. - Progress in understanding the molecular mechanism of Xist RNA function through genetics.
Monfort A, Wutz A. Monfort A, et al. Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160368. doi: 10.1098/rstb.2016.0368. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947663 Free PMC article. Review. - Epigenetic control of chromosome-associated lncRNA genes essential for replication and stability.
Heskett MB, Vouzas AE, Smith LG, Yates PA, Boniface C, Bouhassira EE, Spellman PT, Gilbert DM, Thayer MJ. Heskett MB, et al. Nat Commun. 2022 Oct 22;13(1):6301. doi: 10.1038/s41467-022-34099-7. Nat Commun. 2022. PMID: 36273230 Free PMC article.
References
- Brockdorff N. Curr Opin Genet Dev. 1998;8:328–333. - PubMed
- Hong Y K, Ontiveros S D, Strauss W M. Mamm Genome. 2000;11:220–224. - PubMed
- Memili E, Hong Y K, Kim D H, Ontiveros S D, Strauss W M. Gene. 2001;266:131–137. - PubMed
- Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM45441/GM/NIGMS NIH HHS/United States
- T32 HD007439/HD/NICHD NIH HHS/United States
- R01 GM053234/GM/NIGMS NIH HHS/United States
- GM53234/GM/NIGMS NIH HHS/United States
- HD07439-09/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources