GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis - PubMed (original) (raw)
GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis
Aaron N Chang et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The function of GATA transcription factors in diverse developmental contexts depends in part on physical interaction with cofactors of the Friend of GATA (FOG) family. However, previous studies indicate that FOG-1 may play a GATA-1-independent role in early megakaryopoiesis, suggesting that FOG proteins might act in a GATA factor-independent manner. Here, we have generated mouse knock-in (KI) mutants harboring a critical valine-to-glycine substitution in the amino-terminal zinc fingers of GATA-1 and GATA-2 to ablate FOG interaction. In contrast to male GATA-1(KI) (GATA-1 is located on the X-chromosome) or GATA-2(KI/KI) mice, compound GATA-1(KI) GATA-2(KI/KI) mutant mice display complete megakaryopoietic failure, a phenocopy of FOG-1(-/-) mice. We conclude that FOG-1 requires an interaction with either GATA-1 or -2 as part of its essential role in early megakaryopoiesis. On the basis of these and previous reports, we infer that GATA factor dependence is a critical aspect of FOG protein function.
Figures
Figure 1
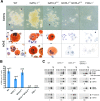
Homologous recombination of targeting constructs and subsequent neo cassette excision in ES cells. (A) Amino acid sequence alignment of the amino-terminal zinc fingers of GATA-1 and GATA-2. Mutated positions in GATA-1 and GATA-2 N-terminal zinc fingers (N-f) are indicated by the red “V” and residue number. Identical sequences are shown in black, and nonidentical residues are shown in blue. (B) Schematic representation of V205G KI mutation gene-targeting strategy into the GATA-1 locus on the X-chromosome. Subsequent Cre-mediated excision of neomycin resistance gene (neo) cassette is shown below. Solid triangles represent loxP sites. Location of probe used for Southern analysis is indicated by solid bar (probe A). PCR primers are shown as horizontally opposed arrows. (C) Southern blot and PCR analyses depicting correctly recombined and Cre-excised GATA-1KI loci. (D) Schematic representation of V296G KI targeting strategy in the GATA-2 locus and subsequent Cre-mediated excision of neo cassette. Locations of probes used for Southern analysis are indicated by solid bars (probes B and C). (E) Southern blot analysis depicting correctly recombined and Cre-excised GATA-2KI loci.
Figure 2
May–Grunwald–Giemsa stains of cytospun erythrocytes collected from E10.5 yolk sacs (original magnification ×1,000).
Figure 3
In vitro megakaryocyte differentiation from E10.5 yolk sac. (A) Phase-contrast appearance and AChE stains of representative megakaryocyte colonies at day 7 of culture. Arrows indicate proplatelet processes. AchE-positive cells stain orange [original magnification, ×320 (phase-contrast); ×1,000 (AchE)]. (B) Quantitation of megakaryocyte colony formation. Colonies were counted on day 7 of culture and expressed as the mean number of megakaryocyte colonies (MK) per 1 × 105cells plated. Error bars represent the SEM. n = number of embryos analyzed. (C) RT-PCR analysis of the murine megakaryocytic markers AChE, von Willebrand factor (vWF), GPIbα and P-selectin, from yolk sac cells cultured for 7 days in liquid culture in the presence of recombinant Tpo. Hypoxanthine ribosyltransferase (HPRT) represents “housekeeping” control gene for normalization of cDNA content. Fold decrease of mRNA levels relative to wild type (WT) (after normalization to HPRT signal) is indicated below each panel. “−RT” indicates no reverse transcriptase was added to sample.
Figure 4
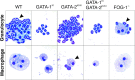
Myeloid colony formation from E10.5 yolk sacs. May–Grunwald–Giemsa stains of cytospun preparation from E10.5 yolk sac cells cultured in methylcellulose in the presence of erythropoietin, Tpo, and Kit ligand for 7 days.
Figure 5
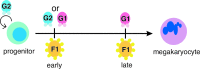
Model of FOG-1–GATA factor interaction requirements during megakaryocyte development. During an early stage of megakaryocyte commitment, FOG-1 requires an interaction with either GATA-1 or GATA-2. During a later stage, FOG-1 specifically requires an interaction with GATA-1. GATA-2 also plays a role in proliferation of progenitor cells (indicated by looping arrow). F1, FOG-1; G1, GATA-1; G2, GATA-2.
Similar articles
- Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors.
Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH. Crispino JD, et al. Genes Dev. 2001 Apr 1;15(7):839-44. doi: 10.1101/gad.875201. Genes Dev. 2001. PMID: 11297508 Free PMC article. - FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation.
Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. Tsang AP, et al. Cell. 1997 Jul 11;90(1):109-19. doi: 10.1016/s0092-8674(00)80318-9. Cell. 1997. PMID: 9230307 - GATA-1: friends, brothers, and coworkers.
Morceau F, Schnekenburger M, Dicato M, Diederich M. Morceau F, et al. Ann N Y Acad Sci. 2004 Dec;1030:537-54. doi: 10.1196/annals.1329.064. Ann N Y Acad Sci. 2004. PMID: 15659837 Review. - In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis.
Shimizu R, Takahashi S, Ohneda K, Engel JD, Yamamoto M. Shimizu R, et al. EMBO J. 2001 Sep 17;20(18):5250-60. doi: 10.1093/emboj/20.18.5250. EMBO J. 2001. PMID: 11566888 Free PMC article. - Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins.
Cantor AB, Orkin SH. Cantor AB, et al. Semin Cell Dev Biol. 2005 Feb;16(1):117-28. doi: 10.1016/j.semcdb.2004.10.006. Epub 2004 Dec 15. Semin Cell Dev Biol. 2005. PMID: 15659346 Review.
Cited by
- NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development.
Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA. Miccio A, et al. EMBO J. 2010 Jan 20;29(2):442-56. doi: 10.1038/emboj.2009.336. Epub 2009 Nov 19. EMBO J. 2010. PMID: 19927129 Free PMC article. - Endothelial lineage-mediated loss of the GATA cofactor Friend of GATA 1 impairs cardiac development.
Katz SG, Williams A, Yang J, Fujiwara Y, Tsang AP, Epstein JA, Orkin SH. Katz SG, et al. Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14030-5. doi: 10.1073/pnas.1936250100. Epub 2003 Nov 12. Proc Natl Acad Sci U S A. 2003. PMID: 14614148 Free PMC article. - Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer.
Huilgol D, Venkataramani P, Nandi S, Bhattacharjee S. Huilgol D, et al. Genes (Basel). 2019 Oct 12;10(10):794. doi: 10.3390/genes10100794. Genes (Basel). 2019. PMID: 31614829 Free PMC article. Review. - The Pleiotropic Effects of GATA1 and KLF1 in Physiological Erythropoiesis and in Dyserythropoietic Disorders.
Barbarani G, Fugazza C, Strouboulis J, Ronchi AE. Barbarani G, et al. Front Physiol. 2019 Feb 12;10:91. doi: 10.3389/fphys.2019.00091. eCollection 2019. Front Physiol. 2019. PMID: 30809156 Free PMC article. Review. - Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development.
Woo AJ, Moran TB, Schindler YL, Choe SK, Langer NB, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB. Woo AJ, et al. Mol Cell Biol. 2008 Apr;28(8):2675-89. doi: 10.1128/MCB.01945-07. Epub 2008 Feb 4. Mol Cell Biol. 2008. PMID: 18250154 Free PMC article.
References
- Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases