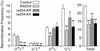Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54 - PubMed (original) (raw)
Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54
Perry M Kim et al. Nucleic Acids Res. 2002.
Abstract
Rad54 plays key roles in homologous recombination (HR) and double-strand break (DSB) repair in yeast, along with Rad51, Rad52, Rad55 and Rad57. Rad54 belongs to the Swi2/Snf2 family of DNA-stimulated ATPases. Rad51 nucleoprotein filaments catalyze DNA strand exchange and Rad54 augments this activity of Rad51. Mutations in the Rad54 ATPase domain (ATPase(-)) impair Rad54 function in vitro, sensitize yeast to killing by methylmethane sulfonate and reduce spontaneous gene conversion. We found that overexpression of ATPase(-) Rad54 reduced spontaneous direct repeat gene conversion and increased both spontaneous direct repeat deletion and spontaneous allelic conversion. Overexpression of ATPase(-) Rad54 decreased DSB-induced allelic conversion, but increased chromosome loss and DSB-dependent lethality. Thus, ATP hydrolysis by Rad54 contributes to genome stability by promoting high-fidelity DSB repair and suppressing spontaneous deletions. Overexpression of wild-type Rad54 did not alter DSB-induced HR levels, but conversion tract lengths were reduced. Interestingly, ATPase(-) Rad54 decreased overall HR levels and increased tract lengths. These tract length changes provide new in vivo evidence that Rad54 functions in the post-synaptic phase during recombinational repair of DSBs.
Figures
Figure 1
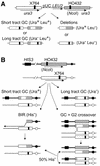
Recombination substrates and products. (A) In haploid strains, 1.2 kb ura3 direct repeats flank pUC19 and LEU2. The left copy is inactivated by X764, and the right copy (shaded) is inactivated by insertion of an HO site at _Nco_I (HO432). The four principal HR products and phenotypes are shown below. Gene conversions (GC) retain LEU2 (Leu+); short tract GC does not convert the X764 frameshift mutation; long tract GC includes X764, giving Ura– products. (B) The same ura3 genes present at allelic positions in diploids. The chromosome carrying the HO site is marked with HIS3 near the chromosome V telomere ∼100 kb from ura3. Conversions not associated with a crossover retain HIS3 (top two products); BIR and 50% of crossovers result in HIS3 loss. Loss of the broken chromosome (not shown) also leads to HIS3 loss.
Figure 2
Immunoblot analysis of wild-type and ATPase– Rad54. Protein extracts were prepared from JW3082 expressing wild-type levels of Rad54 (control), and from JW3082 overexpressing RAD54, rad54-KA or rad54-KR. In each lane 50 µg of protein was loaded, the proteins were separated by SDS–PAGE, transferred to PVDF membranes and detected with antibodies to Rad54 (α-Rad54) using ECL reagents (Amersham Pharmacia Biotech, Piscataway, NJ). α-Rad54 detects both wild-type and ATPase– Rad54. Parallel lanes were probed with antibodies to 3-phosphoglycerate kinase (α-3PGK). A brief exposure is shown to compare overexpression levels; with this exposure Rad54 expressed from the native chromosomal locus cannot be detected.
Figure 3
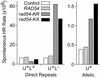
ATPase– Rad54 enhances rates of spontaneous direct repeat deletion and allelic gene conversion. Spontaneous HR yielding Ura+ recombinants was assayed by fluctuation analysis of 11 independent parental colonies of JW3084 (Direct Repeats) or JC3520 (Allelic). For direct repeats, Ura+ products were replica plated to leucine omission medium to determine rates for Ura+ Leu+ (gene conversion) and Ura+ Leu– (deletion). Cells carried an empty (TRP1) vector as control, or they overexpressed wild-type (RAD54) or ATPase– Rad54 (rad54-KR and rad54-KA).
Figure 4
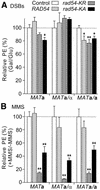
Dominant negative effects of excess wild-type or ATPase– Rad54 on cell survival following DNA damage. (A) DSB-dependent cell killing. PEs were determined following 24 h growth in glycerol medium and 6 h growth in either glucose or galactose medium. DSB-dependent cell killing was determined by dividing the galactose PE by the glucose PE for each of three to four determinations per strain. The ratios were converted to percentages, and the averages ± SEM were plotted after normalizing control values to 100%. Values <100% indicate greater DSB-dependent cell killing relative to control; *, P < 0.05; **, P = 0.012. (B) MMS-dependent cell killing. PEs were determined for colonies grown in the presence or absence of 0.01% MMS for 6 days. MMS-dependent cell killing was determined as the ratio of PEs for MMS-treated to untreated cells for each of four determinations per strain. The ratios were converted to percentages, and the averages ± SEM are plotted after normalizing control values to 100%. Values <100% indicate greater MMS-dependent cell killing relative to control; **, P ≤ 0.01.
Figure 5
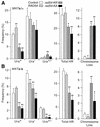
Dominant negative effects of excess wild-type or ATPase– Rad54 on DSB-induced direct repeat recombination. Frequencies of each of the four phenotypic classes, plus totals of all classes are shown for JW3082 carrying a control vector or overexpressing RAD54, rad54-KR or rad54-KA. Data are averages ± SD for four determinations per strain; *, P < 0.02; **, P < 0.003.
Figure 6
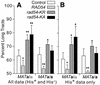
Dominant negative effects of excess wild-type or ATPase– Rad54 on DSB-induced allelic recombination. (A) Frequencies of Ura+, Ura– and sectored Ura+/– products, HR totals and chromosome loss are shown for JC3517-13 (MATa/α) carrying a control vector or overexpressing RAD54, rad54-KR or rad54-KA (see Table 2 for distributions of His+ and His– phenotypes). Data are averages ± SD for four determinations per strain; *, P < 0.05; **, P < 0.01. (B) Data for JC3545 (MATa/a) as above.
Figure 7
Effects of excess wild-type or ATPase– Rad54 on gene conversion tract lengths. (A) Allelic tract lengths for JC3517-13 (MATa/α) and JC3545 (MATa/a) were calculated as the ratio of long tracts (sum of Ura– + half of Ura+/–) divided by total recombinants. Data are averages ± SD for four determinations per strain; *, P ≤ 0.05; **, P < 0.01. (B) Allelic tract lengths calculated from His+ data.
Similar articles
- Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1.
Pohl TJ, Nickoloff JA. Pohl TJ, et al. Mol Cell Biol. 2008 Feb;28(3):897-906. doi: 10.1128/MCB.00524-07. Epub 2007 Nov 26. Mol Cell Biol. 2008. PMID: 18039855 Free PMC article. - Overexpression of Rad51 inhibits double-strand break-induced homologous recombination but does not affect gene conversion tract lengths.
Paffett KS, Clikeman JA, Palmer S, Nickoloff JA. Paffett KS, et al. DNA Repair (Amst). 2005 Jun 8;4(6):687-98. doi: 10.1016/j.dnarep.2005.03.003. DNA Repair (Amst). 2005. PMID: 15878310 - Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament.
Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Solinger JA, et al. J Mol Biol. 2001 Apr 13;307(5):1207-21. doi: 10.1006/jmbi.2001.4555. J Mol Biol. 2001. PMID: 11292336 - Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination.
Ceballos SJ, Heyer WD. Ceballos SJ, et al. Biochim Biophys Acta. 2011 Sep;1809(9):509-23. doi: 10.1016/j.bbagrm.2011.06.006. Epub 2011 Jun 16. Biochim Biophys Acta. 2011. PMID: 21704205 Free PMC article. Review. - Rad54: the Swiss Army knife of homologous recombination?
Heyer WD, Li X, Rolfsmeier M, Zhang XP. Heyer WD, et al. Nucleic Acids Res. 2006;34(15):4115-25. doi: 10.1093/nar/gkl481. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935872 Free PMC article. Review.
Cited by
- Bridge-induced chromosome translocation in yeast relies upon a Rad54/Rdh54-dependent, Pol32-independent pathway.
Tosato V, Sidari S, Bruschi CV. Tosato V, et al. PLoS One. 2013 Apr 17;8(4):e60926. doi: 10.1371/journal.pone.0060926. Print 2013. PLoS One. 2013. PMID: 23613757 Free PMC article. - Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics.
Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Li X, et al. Nucleic Acids Res. 2007;35(12):4124-40. doi: 10.1093/nar/gkm412. Epub 2007 Jun 12. Nucleic Acids Res. 2007. PMID: 17567608 Free PMC article. - The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair.
Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. Sun W, et al. Mol Cell. 2008 Oct 10;32(1):118-28. doi: 10.1016/j.molcel.2008.08.024. Mol Cell. 2008. PMID: 18851838 Free PMC article. - Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1.
Pohl TJ, Nickoloff JA. Pohl TJ, et al. Mol Cell Biol. 2008 Feb;28(3):897-906. doi: 10.1128/MCB.00524-07. Epub 2007 Nov 26. Mol Cell Biol. 2008. PMID: 18039855 Free PMC article. - Rad54 and Rdh54 prevent Srs2-mediated disruption of Rad51 presynaptic filaments.
Meir A, Crickard JB, Kwon Y, Sung P, Greene EC. Meir A, et al. Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2113871119. doi: 10.1073/pnas.2113871119. Proc Natl Acad Sci U S A. 2022. PMID: 35042797 Free PMC article.
References
- Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. - PubMed
- Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials