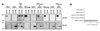FANCE: the link between Fanconi anaemia complex assembly and activity - PubMed (original) (raw)
FANCE: the link between Fanconi anaemia complex assembly and activity
Paul Pace et al. EMBO J. 2002.
Abstract
The Fanconi anaemia (FA) nuclear complex (composed of the FA proteins A, C, G and F) is essential for protection against chromosome breakage. It activates the downstream protein FANCD2 by monoubiquitylation; this then forges an association with the BRCA1 protein at sites of DNA damage. Here we show that the recently identified FANCE protein is part of this nuclear complex, binding both FANCC and FANCD2. Indeed, FANCE is required for the nuclear accumulation of FANCC and provides a critical bridge between the FA complex and FANCD2. Disease-associated FANCC mutants do not bind to FANCE, cannot accumulate in the nucleus and are unable to prevent chromosome breakage.
Figures
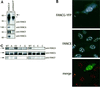
Fig. 1. FANCE is a 58 kDa protein and is localized to nuclear foci. (A and B) Detection and analysis of FANCE protein, either immunoprecipitated with FANCE antiserum or with Flag monoclonal, then blotted with the FANCE antiserum (A) or with Flag monoclonal (B). The FANCE antiserum specifically immunoprecipitates and western blots endogenous FANCE, as well as recombinant FANCE with a C-terminal 2×Flag epitope (FANCE-2×Flag) expressed in HeLa cells. The arrows mark the migration of FANCE and FANCE-2×Flag. (C) Subcellular localization of endogenous FANCE as well as transfected FANCE–YFP, as detected by indirect immunofluoresence. FANCE antiserum detects FANCE–YFP transiently expressed in transfected (yellow arrow), as well as native protein in foci of untransfected (red arrow) HeLa cells. The preimmune serum and second layer (Cy5 anti-rabbit γ chain) gave no signal (data not shown). (D) Whole-cell lysate western blot analysis for FANCE expression with FANCE antiserum in nuclear extract from cell lines comprising the A, C, D2, E, F and G complementation groups. FANCE is present in all groups except, as expected, in the FANCE complementation group.
Fig. 2. FANCE is part of the FA nuclear complex. (A) Western blot analysis of HeLa nuclear extract immunoprecipitated with anti-FANCE rabbit antiserum and protein A beads. Samples were western blotted with rabbit anti-FANCE, -FANCA, -FANCF and -FANCG antiserum. FANCE co-purifies with FANCA, FANCF and FANCG (the asterisk marks the presence of rabbit γ chain). (B) Co-localization in nuclear foci of transiently transfected FA complex protein FANCG–YFP (green) with endogenous FANCE (red) in HeLa cells. Recombinant FANCG–YFP can be seen to localize to the cytoplasm as well as nuclear foci; in contrast, endogenous FANCE is concentrated in nuclear foci. Upon merging of the layers, the FANCE foci co-localize with the signal from transfected FANCG–YFP. FANCE was detected with FANCE antiserum followed by Cy5 anti-rabbit γ chain. (C) FANCE immunoprecipitations in cell lines from the A, C, D2, E, F and G complementation groups were blotted with anti-FANCA, FANCG and FANCF antisera. The interactions between FA complex proteins and FANCE are disrupted in all except the D2 complementation group.
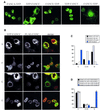
Fig. 3. FANCE is essential for the nuclear accumulation of FANCC. (A) Expression of transfected FA complex proteins (FANCA, FANCC, FANCF, FANCG and FANCE) with either N- or C-terminal YFP tag in COS cells. All the FA complex proteins show strong nuclear accumulation except for YFP–FANCC, which is predominantly in the cytoplasmic compartment. (B) Co-expression of YFP–FANCC (green) with C-terminal Flag-tagged FA complex proteins (FANCA, FANCF, FANCG and FANCE, all in red) in COS cells. Only co-expression of FANCE leads to the nuclear accumulation of FANCC. (C) One hundred transfected cells were scored blind for the localization of the Flag tag as well as YFP–FANCC to cytoplasmic (C), cytoplasmic/nuclear (C+N) and nuclear (N) compartments in transfected COS cells. Only FANCC is predominantly in the cytoplasmic compartment. (D) Localization of YFP–FANCC when co-expressed with Flag-tagged FANCA, FANCF, FANCG or FANCE. The scores are expressed as an average from three independent observers. Only FANCE-Flag co-expression leads to FANCC nuclear accumulation.
Fig. 4. FANCC mutants are defective in binding FANCE. (A) Co-immunoprecipitation of protein C epitope-tagged FANCE with GST-tagged wild-type (WT) or mutant (L496R and L554P) FANCC in baculovirus-infected insect cells. The gel transfers were western blotted with anti-protein C monoclonal antibody (for the detection of FANCE) or with anti-GST antisera (for the detection of FANCC). The interaction between FANCE and WT FANCC is readily detectable, while that with the mutants is reduced. (B) Mammalian two hybrid interaction (M2H) test showing strong induction of the reporter (luciferase) in cells expressing WT FANCC and FANCE but not in the case of mutant FANCC. Units are expressed as fold induction of luciferase reporter when compared with either gene with empty vector (pAM or VP-16) alone.
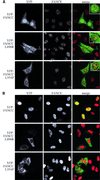
Fig. 5. FANCC mutants do not accumulate in the nucleus. (A) Localization of transiently transfected YFP–FANCC (green) and YFP–FANCC mutants in HeLa cells. Endogenous FANCE is present in nuclear foci (red), co-localizing with WT FANCC but not with mutant FANCC (see insets). (B) As (A), but in HeLa cells that stably overexpress FANCE-2×Flag. Wild-type FANCC accumulates in the nucleus, while neither of the mutants do.
Fig. 6. FANCE binds to and co-localizes with the downstream FA protein FANCD2. (A) M2H test indicating that FANCE, but not any of the other FA complex proteins, interacts with FANCD2 (activation domain-fused FA complex protein tested against DNA-binding domain-fused FANCD2). (B) FANCD2 co-elutes with His12-tagged FANCE on a nickel-NTA matrix when captured from lysates of baculovirus-infected insect cells expressing both proteins. Insect cells expressing only FANCD2 were used as a control. FT, flow-through; W1, first wash; W10, tenth wash; E, elution with imidazole. Gels were transferred and blotted with anti-FANCE antiserum or anti-FANCD2 antisera. (C) Coomassie Blue stain of FANCD2/FANCE-His12 complexes purified by Ni2+–agarose chromatography from baculovirus-infected insect cells expressing both proteins. Both proteins co-purify efficiently. (D) Co-localization of transiently expressed YFP–FANCD2 (green) with endogenous FANCE (red) or endogenous FANCD2 (red) with transiently expressed FANCE–YFP (green), in irradiated (6 h post-3 Gy) HeLa cells. In the left-hand panel, transfected YFP–FANCD2 (top) as well as endogenous FANCE (middle) localize to nuclear foci. Merging of the images shows co-localization for some of these foci. The right-hand panel shows the same experiment, but this time with the tagging combination reversed, i.e. FANCE is now YFP tagged and transfected, and it is endogeneous FANCD2 that is being assayed. Top, transfected FANCE–YFP. Middle, endogenous FANCD2 (detected by rabbit anti-FANCD2 antisera followed by anti-rabbit Cy5 antibody). Bottom, merged image. (E) Endogenous FANCE co-purifies with endogenous FANCD2 in HeLa nuclear extract. HeLa nuclear extract (4 mg) was incubated with protein A–Sepharose beads without antibody (B), with rabbit anti-FANCE antisera (EIP) and with control preimmune antisera (C). The samples were western blotted with anti-FANCD2 antisera (top) and anti-FANCE antisera (bottom). The arrows indicate the migration of FANCD2 and FANCE.
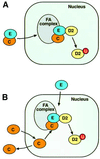
Fig. 7. A model for the assembly of the FA nuclear complex. (A) The FANCE protein carries FANCC into the nucleus and assembles the FA complex. (B) FANCE and FANCC enter the nucleus independently and FANCC is retained in this compartment upon complex assembly.
Similar articles
- Evaluation of Fanconi Anemia genes in familial breast cancer predisposition.
Seal S, Barfoot R, Jayatilake H, Smith P, Renwick A, Bascombe L, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N; Breast Cancer Susceptibility Collaboration. Seal S, et al. Cancer Res. 2003 Dec 15;63(24):8596-9. Cancer Res. 2003. PMID: 14695169 - The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia.
Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, Ott J, Petrini J, Schindler D, Hanenberg H, Auerbach AD. Levran O, et al. Nat Genet. 2005 Sep;37(9):931-3. doi: 10.1038/ng1624. Epub 2005 Aug 21. Nat Genet. 2005. PMID: 16116424 - Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1.
Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG. Hussain S, et al. Hum Mol Genet. 2003 Oct 1;12(19):2503-10. doi: 10.1093/hmg/ddg266. Epub 2003 Aug 5. Hum Mol Genet. 2003. PMID: 12915460 - Molecular biology of Fanconi anaemia--an old problem, a new insight.
Ahmad SI, Hanaoka F, Kirk SH. Ahmad SI, et al. Bioessays. 2002 May;24(5):439-48. doi: 10.1002/bies.10082. Bioessays. 2002. PMID: 12001267 Review. - [Molecular basis of Fanconi's anemia].
Digweed M. Digweed M. Klin Padiatr. 1999 Jul-Aug;211(4):192-7. doi: 10.1055/s-2008-1043786. Klin Padiatr. 1999. PMID: 10472548 Review. German.
Cited by
- A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome.
Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. Meetei AR, et al. Mol Cell Biol. 2003 May;23(10):3417-26. doi: 10.1128/MCB.23.10.3417-3426.2003. Mol Cell Biol. 2003. PMID: 12724401 Free PMC article. - Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells.
Yamamoto K, Ishiai M, Matsushita N, Arakawa H, Lamerdin JE, Buerstedde JM, Tanimoto M, Harada M, Thompson LH, Takata M. Yamamoto K, et al. Mol Cell Biol. 2003 Aug;23(15):5421-30. doi: 10.1128/MCB.23.15.5421-5430.2003. Mol Cell Biol. 2003. PMID: 12861027 Free PMC article. - The Fanconi anemia protein interaction network: casting a wide net.
Rego MA, Kolling FW 4th, Howlett NG. Rego MA, et al. Mutat Res. 2009 Jul 31;668(1-2):27-41. doi: 10.1016/j.mrfmmm.2008.11.018. Epub 2008 Dec 3. Mutat Res. 2009. PMID: 19101576 Free PMC article. Review. - Identification of Three Novel Mutations in the FANCA, FANCC, and ITGA2B Genes by Whole Exome Sequencing.
Negahdari S, Zamani M, Seifi T, Sedighzadeh S, Mazaheri N, Zeighami J, Sedaghat A, Saberi A, Hamid M, Keikhaei B, Radpour R, Shariati G, Galehdari H. Negahdari S, et al. Int J Prev Med. 2020 Aug 6;11:117. doi: 10.4103/ijpvm.IJPVM_462_19. eCollection 2020. Int J Prev Med. 2020. PMID: 33088445 Free PMC article. - How the fanconi anemia pathway guards the genome.
Moldovan GL, D'Andrea AD. Moldovan GL, et al. Annu Rev Genet. 2009;43:223-49. doi: 10.1146/annurev-genet-102108-134222. Annu Rev Genet. 2009. PMID: 19686080 Free PMC article. Review.
References
- Auerbach A.D. and Wolman,S.R. (1976) Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature, 261, 494–496. - PubMed
- Auerbach A.D., Buchwald,M. and Joenje,H. (2001) The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, NY.
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. - PubMed
- Cumming R.C., Lightfoot,J., Beard,K., Youssoufian,H., O’Brien,P.J. and Buchwald,M. (2001) Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat. Med., 7, 814–820. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous