Testing for population subdivision and association in four case-control studies - PubMed (original) (raw)
Testing for population subdivision and association in four case-control studies
Kristin G Ardlie et al. Am J Hum Genet. 2002 Aug.
Abstract
Population structure has been presumed to cause many of the unreplicated disease-marker associations reported in the literature, yet few actual case-control studies have been evaluated for the presence of structure. Here, we examine four moderate case-control samples, comprising 3,472 individuals, to determine if detectable population subdivision is present. The four population samples include: 500 U.S. whites and 236 African Americans with hypertension; and 500 U.S. whites and 500 Polish whites with type 2 diabetes, all with matched control subjects. Both diabetes populations were typed for the PPARg Pro12Ala polymorphism, to replicate this well-supported association (Altshuler et al. 2000). In each of the four samples, we tested for structure, using the sum of the case-control allele frequency chi(2) statistics for 9 STR and 35 SNP markers (Pritchard and Rosenberg 1999). We found weak evidence for population structure in the African American sample only, but further refinement of the sample, to include only individuals with U.S.-born parents and grandparents, eliminated the stratification. Our examples provide insight into the factors affecting the replication of association studies and suggest that carefully matched, moderate-sized case-control samples in cosmopolitan U.S. and European populations are unlikely to contain levels of structure that would result in significantly inflated numbers of false-positive associations. We explore the role that extreme differences in power among studies, due to sample size and risk-allele frequency differences, may play in the replication problem.
Figures
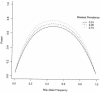
Figure 1
Power to detect an allelic association for 500 case subjects and 500 control subjects under a multiplicative genetic model with genotypic risk ratio 1.25, by risk-allele frequency and disease prevalence. Allele frequencies for case and control subjects were determined under the given genetic models; power to detect the difference in allele frequencies was calculated for the exact test for a 2 × 2 contingency table, by the method of Walters (1979).
Comment in
- Regarding "Testing for population subdivision and association in four case-control studies".
Pankow JS, Province MA, Hunt SC, Arnett DK. Pankow JS, et al. Am J Hum Genet. 2002 Dec;71(6):1478-80. doi: 10.1086/344582. Am J Hum Genet. 2002. PMID: 12515254 Free PMC article. No abstract available.
Similar articles
- Is a Pro12Ala polymorphism of the PPARgamma2 gene related to obesity and type 2 diabetes mellitus in the Czech population?
Srámková D, Kunesová M, Hainer V, Hill M, Vcelák J, Bendlová B. Srámková D, et al. Ann N Y Acad Sci. 2002 Jun;967:265-73. doi: 10.1111/j.1749-6632.2002.tb04282.x. Ann N Y Acad Sci. 2002. PMID: 12079854 - The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes.
Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. Altshuler D, et al. Nat Genet. 2000 Sep;26(1):76-80. doi: 10.1038/79216. Nat Genet. 2000. PMID: 10973253 - [Association of Pro12Ala variant in peroxisome proliferator-activated receptor-gamma2 gene with type 2 diabetes mellitus].
Fu M, Chen H, Li X, Li J, Wu B, Cheng L, Cai M, Fu Z. Fu M, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002 Jun;19(3):234-8. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002. PMID: 12048686 Chinese. - Assessing the impact of population stratification on genetic association studies.
Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Freedman ML, et al. Nat Genet. 2004 Apr;36(4):388-93. doi: 10.1038/ng1333. Epub 2004 Mar 28. Nat Genet. 2004. PMID: 15052270 - The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism.
Stumvoll M, Häring H. Stumvoll M, et al. Diabetes. 2002 Aug;51(8):2341-7. doi: 10.2337/diabetes.51.8.2341. Diabetes. 2002. PMID: 12145143 Review.
Cited by
- Population stratification confounds genetic association studies among Latinos.
Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG; Genetics of Asthma in Latino Americans GALA Study. Choudhry S, et al. Hum Genet. 2006 Jan;118(5):652-64. doi: 10.1007/s00439-005-0071-3. Epub 2005 Nov 8. Hum Genet. 2006. PMID: 16283388 - Common variants at the PCOL2 and Sp1 binding sites of the COL1A1 gene and their interactive effect influence bone mineral density in Caucasians.
Liu PY, Lu Y, Long JR, Xu FH, Shen H, Recker RR, Deng HW. Liu PY, et al. J Med Genet. 2004 Oct;41(10):752-7. doi: 10.1136/jmg.2004.019851. J Med Genet. 2004. PMID: 15466008 Free PMC article. - Genetic nurturing, missing heritability, and causal analysis in genetic statistics.
Shen H, Feldman MW. Shen H, et al. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25646-25654. doi: 10.1073/pnas.2015869117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989124 Free PMC article. - Posterior predictive checks to quantify lack-of-fit in admixture models of latent population structure.
Mimno D, Blei DM, Engelhardt BE. Mimno D, et al. Proc Natl Acad Sci U S A. 2015 Jun 30;112(26):E3441-50. doi: 10.1073/pnas.1412301112. Epub 2015 Jun 12. Proc Natl Acad Sci U S A. 2015. PMID: 26071445 Free PMC article. - Association of the DTNBP1 locus with schizophrenia in a U.S. population.
Funke B, Finn CT, Plocik AM, Lake S, DeRosse P, Kane JM, Kucherlapati R, Malhotra AK. Funke B, et al. Am J Hum Genet. 2004 Nov;75(5):891-8. doi: 10.1086/425279. Epub 2004 Sep 10. Am J Hum Genet. 2004. PMID: 15362017 Free PMC article.
References
Electronic-Database Information
- Global Repository at GCI, http://www.genomicsinc.com (for case-control samples)
- SNP Consortium, http://snp.cshl.org/allele_frequency_project/ (for biallelic SNPs)
- World Health Organization, http://www.who.int/ncd/dia/databases2.htm#t3
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARg Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 - PubMed
- Breslow NE, Day NE (1994) Statistical methods in cancer research, volume II: the design and analysis of cohort studies. IARC Scientific Publications, No. 82, Oxford University Press, New York - PubMed
- Destro-Bisol G, Maviglia R, Caglia A, Boschi I, Spedini G, Pascali V, Clark A, Tishkoff S (1999) Estimating European admixture in African Americans by using microsatellites and a microsatellite haplotype (CD4/Alu). Hum Genet 104:149–157 - PubMed
- Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources