Two different mechanisms of disinhibition produced by GABAA receptor mutations linked to epilepsy in humans - PubMed (original) (raw)
Two different mechanisms of disinhibition produced by GABAA receptor mutations linked to epilepsy in humans
Matt T Bianchi et al. J Neurosci. 2002.
Abstract
The first mutations of the GABA(A) receptor channel linked to familial epilepsy in humans were reported recently (Baulac et al., 2001; Wallace et al., 2001). Preliminary functional analysis of alpha1beta2gamma2 GABA(A) receptors expressed in Xenopus oocytes suggested that the gamma2 subunit R43Q mutation abolished current enhancement by the benzodiazepine, diazepam, and that the gamma2 subunit K289M mutation decreased current amplitudes. We used single-channel recording and concentration jump techniques applied to outside out patches to evaluate the impact of these mutations on GABA(A) receptor channel function of the highly conserved rat ortholog subunits expressed in human embryonic kidney cells. When coexpressed with alpha1 and beta3 subunits, no differences were observed between wild-type and mutant GABA(A) receptor current activation rates or rates or extent of desensitization during prolonged (400 msec) GABA application (1 mm). Although deactivation after brief (5 msec) or prolonged (400 msec) GABA application was unaltered by the R43Q mutation, deactivation (a correlate of IPSC duration) was accelerated for the K289M mutation. Faster deactivation was likely a consequence of altered gating, because single-channel openings had shorter mean duration. Interestingly, the R43Q mutation did not alter diazepam potentiation. It did, however, substantially decrease current amplitude, which was not caused by decreased single-channel conductance or open time, suggesting reduced surface expression of functional receptors. The two gamma2 subunit mutations likely produce disinhibition and familial epilepsy by distinct mechanisms, suggesting that maintenance of neuronal inhibition depends not only on the peak amplitude of IPSCs, but also on their time course.
Figures
Fig. 1.
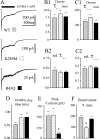
Sensitivity to GABA and diazepam.A, The EC50 for GABA was determined by whole-cell responses to increasing GABA concentration, normalized to the peak current amplitude in each cell. Cells were voltage clamped at –10 to –50 mV, and data were obtained from three cells for each mutation. The dotted line is the concentration–response of wild-type α1β3γ2L GABAAreceptors (Bianchi et al., 2001). B, Diazepam sensitivity was determined by coapplication of 1 μ
m
diazepam with 2 μ
m
GABA (∼EC20) to whole cells expressing α1β3γ2L (top), α1β3γ2L(K289M), or α1β3γ2L(R43Q) GABAAreceptors. C, Diazepam enhancement is shown as the percentage of control responses (averagepeak current before and after diazepam coapplication). Neither mutation significantly affected modulation of GABA-evoked currents by diazepam. D, Representative currents are shown for two human α1β3γ2L(R43Q) GABAA receptor isoforms. The left pair of traces indicated the response of receptors containing a γ2L subunit with amino acid sequence corresponding to the original published γ2L sequence, whereas the_right pair_ of traces indicated the response of receptors containing a γ2L subunit with amino acid sequence corresponding to that of the rat γ2L subunit (see Materials and Methods). For both pairs of traces, the left current was evoked with 10 μ
m
GABA (solid bar), and the right current was evoked with coapplied 1 μ
m
diazepam (hatched bar) in the same cell. Similar results were obtained in five cells.
Fig. 2.
Macroscopic kinetic properties. A, Representative current traces obtained from wild-type or mutated receptors during 400 msec jumps into 1 m
m
GABA. Time scale of top trace applies to all three traces.B1_–_C2, Neither the fast (B1) nor the slow (C1) time constant of desensitization, nor their relative contributions (B2,C2) were significantly altered by the mutations.D, Current activation rate, as indicated by the 10–90% rise time of the current, was not significantly altered by the mutations. E, Peak current amplitudes were significantly smaller for α1β3γ2L(R43Q) GABAA receptors. *p < 0.01. F, Current deactivation after removal of GABA was significantly faster for α1β3γ2L(K289M) GABAA receptors. *p < 0.001. Data were obtained from 8–13 patches.
Fig. 3.
Deactivation after brief GABA pulses.A, Representative currents illustrate deactivation rates of α1β3γ2L (A1), α1β3γ2L(K289M) (A2), and α1β3γ2L(R43Q) (A3) GABAA receptors in response to brief (<5 msec) pulses of GABA (1 m
m
). Scale bars apply to all three_solid traces_. The solid trace in_A3_ is expanded 10-fold (gray trace) for comparison of deactivation current time course.B, Weighted time constants of deactivation (see Materials and Methods) are shown for wild-type and mutated channels. Deactivation was significantly faster for α1β3γ2L(K289M) GABAA receptors (hatched bar). *p < 0.05. α1β3γ2L (R43Q) GABAAreceptor deactivation (solid bar) was not different than that of wild-type receptors (gray bar). Data were obtained from 9–13 patches for each isoform.
Fig. 4.
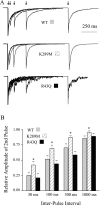
Paired pulse inhibition. Pairs of 5 msec GABA (1 m
m
) pulses were delivered to outside-out patches at interpulse intervals of 30, 100, 300, and 1000 msec. A, Representative currents from wild-type and mutated GABAAreceptors. B, Summary plot showing the relative amplitude of the second pulse of each pair, for each four interpulse intervals. Less inhibition was observed for α1β3γ2L(K289M) GABAA receptors for each interpulse interval.
Fig. 5.
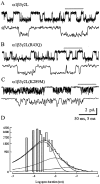
Single-channel analysis. Single-channel records obtained in patches held at −75 mV in the presence of 1 m
m
GABA from α1β3γ2L (A), α1β3γ2L(R43Q) (B), and α1β3γ2L(K289M) (C) GABAA receptors. A portion of the_top trace_ in each pair (indicated by the open bar) is expanded below that trace. Calibration bars apply to all three panels. Openings are downward. Similar results were observed from three α1β3γ2L, five α1β3γ2L(R43Q), and seven α1β3γ2L(K289M) patches. D, Open duration histogram for α1β3γ2L(K289M) single channels. The distribution was best described by the sum of three exponential functions, with each exponential fit shown as a smooth curve. Although three functions were required, the relative contribution of the shortest open state (leftmost curve) was highest, indicating that most openings were brief in duration. The time constants were 0.33, 1.17, and 4.54 msec with relative areas 0.81, 0.17, and 0.02, respectively. Data were pooled from seven α1β3γ2L(K289M) patches.
Similar articles
- mRNA surveillance and endoplasmic reticulum quality control processes alter biogenesis of mutant GABAA receptor subunits associated with genetic epilepsies.
Macdonald RL, Kang JQ. Macdonald RL, et al. Epilepsia. 2012 Dec;53 Suppl 9(0 9):59-70. doi: 10.1111/epi.12035. Epilepsia. 2012. PMID: 23216579 Free PMC article. Review. - Mutations linked to generalized epilepsy in humans reduce GABA(A) receptor current.
Macdonald RL, Bianchi MT, Feng H. Macdonald RL, et al. Exp Neurol. 2003 Nov;184 Suppl 1:S58-67. doi: 10.1016/j.expneurol.2003.08.011. Exp Neurol. 2003. PMID: 14597328 - Altered kinetics and benzodiazepine sensitivity of a GABAA receptor subunit mutation [gamma 2(R43Q)] found in human epilepsy.
Bowser DN, Wagner DA, Czajkowski C, Cromer BA, Parker MW, Wallace RH, Harkin LA, Mulley JC, Marini C, Berkovic SF, Williams DA, Jones MV, Petrou S. Bowser DN, et al. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):15170-5. doi: 10.1073/pnas.212320199. Epub 2002 Nov 1. Proc Natl Acad Sci U S A. 2002. PMID: 12415111 Free PMC article. - Molecular Pathogenic Basis for GABRG2 Mutations Associated With a Spectrum of Epilepsy Syndromes, From Generalized Absence Epilepsy to Dravet Syndrome.
Kang JQ, Macdonald RL. Kang JQ, et al. JAMA Neurol. 2016 Aug 1;73(8):1009-16. doi: 10.1001/jamaneurol.2016.0449. JAMA Neurol. 2016. PMID: 27367160 Free PMC article. Review.
Cited by
- Electrophysiology of ionotropic GABA receptors.
Sallard E, Letourneur D, Legendre P. Sallard E, et al. Cell Mol Life Sci. 2021 Jul;78(13):5341-5370. doi: 10.1007/s00018-021-03846-2. Epub 2021 Jun 1. Cell Mol Life Sci. 2021. PMID: 34061215 Free PMC article. Review. - GABA(A) receptor gamma 2 subunit mutations linked to human epileptic syndromes differentially affect phasic and tonic inhibition.
Eugène E, Depienne C, Baulac S, Baulac M, Fritschy JM, Le Guern E, Miles R, Poncer JC. Eugène E, et al. J Neurosci. 2007 Dec 19;27(51):14108-16. doi: 10.1523/JNEUROSCI.2618-07.2007. J Neurosci. 2007. PMID: 18094250 Free PMC article. - Neocortical post-traumatic epileptogenesis is associated with loss of GABAergic neurons.
Avramescu S, Nita DA, Timofeev I. Avramescu S, et al. J Neurotrauma. 2009 May;26(5):799-812. doi: 10.1089/neu.2008.0739. J Neurotrauma. 2009. PMID: 19422294 Free PMC article. - mRNA surveillance and endoplasmic reticulum quality control processes alter biogenesis of mutant GABAA receptor subunits associated with genetic epilepsies.
Macdonald RL, Kang JQ. Macdonald RL, et al. Epilepsia. 2012 Dec;53 Suppl 9(0 9):59-70. doi: 10.1111/epi.12035. Epilepsia. 2012. PMID: 23216579 Free PMC article. Review. - The double whammy of ER-retention and dominant-negative effects in numerous autosomal dominant diseases: significance in disease mechanisms and therapy.
Gariballa N, Mohamed F, Badawi S, Ali BR. Gariballa N, et al. J Biomed Sci. 2024 Jun 27;31(1):64. doi: 10.1186/s12929-024-01054-1. J Biomed Sci. 2024. PMID: 38937821 Free PMC article. Review.
References
- Bianchi MT, Macdonald RL. Mutation of the 9′ leucine in the GABAA receptor γ2L subunit produces an apparent decrease in desensitization by stabilizing open states without altering desensitized states. Neuropharmacology. 2001b;41:737–744. - PubMed
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud'homee J-F, Baulac M, Brice A, Bruzzone R, LeGuern E. First genetic evidence of GABAA receptor dysfunction in epilepsy: a mutation in the γ2-subunit gene. Nat Genet. 2001;28:46–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS033300/NS/NINDS NIH HHS/United States
- T32 DA007281/DA/NIDA NIH HHS/United States
- R01-NS33300/NS/NINDS NIH HHS/United States
- T32-DA07281-03/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources