Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles - PubMed (original) (raw)
Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles
Reginald F Clayton et al. J Virol. 2002 Aug.
Erratum in
- J Virol 2002 Sep;76(18):9562
Abstract
Purification of hepatitis C virus (HCV) from sera of infected patients has proven elusive, hampering efforts to perform structure-function analysis of the viral components. Recombinant forms of the viral glycoproteins have been used instead for functional studies, but uncertainty exists as to whether they closely mimic the virion proteins. Here, we used HCV virus-like particles (VLPs) generated in insect cells infected with a recombinant baculovirus expressing viral structural proteins. Electron microscopic analysis revealed a population of pleomorphic VLPs that were at least partially enveloped with bilayer membranes and had viral glycoprotein spikes protruding from the surface. Immunogold labeling using specific monoclonal antibodies (MAbs) demonstrated these protrusions to be the E1 and E2 glycoproteins. A panel of anti-E2 MAbs was used to probe the surface topology of E2 on the VLPs and to compare the antigenicity of the VLPs with that of truncated E2 (E2(660)) or the full-length (FL) E1E2 complex expressed in mammalian cells. While most MAbs bound to all forms of antigen, a number of others showed striking differences in their abilities to recognize the various E2 forms. All MAbs directed against hypervariable region 1 (HVR-1) recognized both native and denatured E2(660) with comparable affinities, but most bound either weakly or not at all to the FL E1E2 complex or to VLPs. HVR-1 on VLPs was accessible to these MAbs only after denaturation. Importantly, a subset of MAbs specific for amino acids 464 to 475 and 524 to 535 recognized E2(660) but not VLPs or FL E1E2 complex. The antigenic differences between E2(660,) FL E1E2, and VLPs strongly point to the existence of structural differences, which may have functional relevance. Trypsin treatment of VLPs removed the N-terminal part of E2, resulting in a 42-kDa fragment. In the presence of detergent, this was further reduced to a trypsin-resistant 25-kDa fragment, which could be useful for structural studies.
Figures
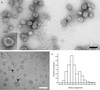
FIG. 1.
(a) Negatively stained electron micrograph of HCV VLPs. (Inset) Higher-magnification image of a VLP that is angular in appearance, with many spikes that appear Y-shaped. (b) Cryomicrograph of HCV VLPs, showing the presence of a lipid bilayer (arrowheads), with protruding spikes, surrounding the core structure. Bars, 100 nm. (c) Histogram of radii measured from 248 VLPs imaged by electron cryomicroscopy.
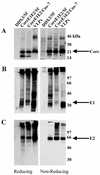
FIG. 2.
Western immunoblot analysis of HCV structural proteins. Extracts of Sf cells infected with rbac-DDX3 or rbac-B45, extracts of COS-7 cells infected with recombinant vaccinia virus v1-836, and VLPs were subjected to SDS-10% PAGE under reducing and nonreducing conditions. Fractionated proteins were immunoblotted with anti-core antiserum R526 (A), anti-E1 antiserum R528 (B), and anti-E2 MAb AP33 (C). Positions of protein size markers are shown.
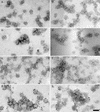
FIG. 3.
Immunogold labeling of HCV VLPs. VLPs placed on EM grids were incubated either singly with MAbs specific for E1 (a) or E2 (b to f), with the polyclonal anti-E2 antiserum R646 (g), or doubly with the anti-E1 serum R528 and the anti-E2 MAb AP33 (h). Anti-mouse IgG and anti-rabbit IgG conjugated with 5- or 10-nm gold particles, respectively, were used as secondary reagents. Grids were washed, stained, and examined under the EM. Bar, 100 nm.
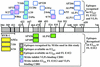
FIG. 4.
Surface topology of E2 epitopes exposed on VLPs. This map was constructed mainly on the basis of the data presented in Table 1, and also on the basis of recently published findings which identified some MAbs as inhibitors of E2-CD81 interaction (42).
FIG. 5.
Trypsin treatment of HCV VLPs. VLPs were either left untreated or treated with trypsin in the presence or absence of Triton X-100. Products were fractionated by SDS-PAGE, and proteins were detected by using the anti-E2 MAbs shown. Positions of molecular weight markers are indicated.
FIG. 6.
Amino acid sequence of HCV E2 (aa 384 to 746 of the HCV polyprotein). Potential trypsin cleavage sites are boldfaced and underlined. Solid bars below amino acid residues, predicted N-linked glycosylation sites. Shaded boxes, epitopes recognized by MAbs AP33, 6/53, and ALP98. The sequence of the TMD (aa 718 to 746) is italicized.
References
- Al-Khayat, H. A., D. Bhella, J. M. Kenney, J. F. Roth, A. J. Kingsman, E. Martin-Rendon, and H. R. Saibil. 1999. Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J. Mol. Biol. 292**:**65-73. -PubMed
- Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117**:**1397-1407. -PubMed
- Bielefeldt Ohmann, H., and B. Bloch. 1982. Electron microscopic studies of bovine viral diarrhea virus in tissues of diseased calves and in cell cultures. Arch. Virol. 71**:**57-74. -PubMed
- Bishop, D. H. L. 1992. Baculovirus expression vectors. Semin. Virol. 3**:**253-264.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources