Negative regulation of transcription by the type II arginine methyltransferase PRMT5 - PubMed (original) (raw)
Negative regulation of transcription by the type II arginine methyltransferase PRMT5
Eric Fabbrizio et al. EMBO Rep. 2002 Jul.
Abstract
We have identified previously a repressor element in the transcription start site region of the cyclin E1 promoter that periodically associates with an atypical, high molecular weight E2F complex, termed CERC. Purification of native CERC reveals the presence of the type II arginine methyltransferase PRMT5, which can mono- or symetrically dimethylate arginine residues in proteins. Chromatin immunoprecipitations (ChIPs) show that PRMT5 is associated specifically with the transcription start site region of the cyclin E1 promoter. ChIP analyses also show that this correlates with the presence on the same promoter region of arginine-methylated proteins including histone H4, an in vitro substrate of PRMT5. Consistent with its presence within the repressor complex, forced expression of PRMT5 negatively affects cyclin E1 promoter activity and cellular proliferation, effects that require its methyltransferase activity. These data provide the first direct experimental evidence that a type II arginine methylase is involved in the control of transcription and proliferation.
Figures
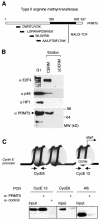
Fig. 1. The type II arginine methyltransferase, PRMT5, is present in CERC. (A) PRMT5 is a component of the native CERC complex; MALDI-MS analysis of affinity purified CERC. Monoisotopic peptides corresponding to PRMT5 are indicated. (B) Western blot analyses of proteins eluted from the CERM affinity column or from the control column (ΔCERM) confirm the presence of PRMT5 in native CERC. Membranes were probed with various antisera as indicated. (C) ChIPs show that PRMT5 is present on the cyclin E1 promoter in vivo. Formaldehyde crosslinked chromatin from G0-arrested NIH 3T3 cells was immunoprecipitated with PRMT5 antisera or control antibody (anti-Flag epitope). One-tenth of the input chromatin and the immunoprecipitated chromatin were analysed by PCR for mouse cyclin E1 promoter fragments (CE12, centred around the transcription start site region and encompassing the CERM element: –40, +46; CE6, centred 500 bp upstream in the promoter region: –432, –526) or the mouse albumin gene. These cyclin E1 promoter DNA fragments are present in two distinct nucleosomes.
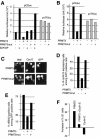
Fig. 2. PRMT5 has an inhibitory effect on cyclin E1 gene transcription and cell proliferation. (A) NIH 3T3 cells were transfected with luciferase reporter genes driven by either the mouse cyclin E1 promoter (pCEluc) or the HSV TK promoter (pvTKluc), together with combinations of expression vectors encoding E2F1, DP1, PRMT5 or an enzymatically inactive PRMT5mut, as indicated. All were co-transfected with a CMV–β-gal reporter construct. Results are expressed in relative luciferase units (RLU), normalised to β-gal activity. (B) Xenopus oocytes were injected with pCEluc or pvTKluc, together with expression vectors encoding either PRMT5 or PRMT5mut, as indicated. Luciferase activities were normalised to β-gal values. (C) NIH 3T3 cells grown on coverslips were transfected with PRMT5 or PRMT5mut expression vectors together with a CMV-driven GFP expression plasmid used to identify transfected cells. Twenty-four hours later, cells were analysed for endogenous cyclin E1 expression by immunofluorescence. Panels show Hoechst staining and cyclin E1 immunodetection in GFP-positive cells. (D) Quantification of several experiments performed as described in (C). Results are expressed as the percentage of transfected cells (GFP positive) that expressed endogenous cyclin E1 protein. (E) Forced expression of PRMT5 blocks cells in G1. PRMT5/GFP co-transfectants were synchronised in G0 by serum depletion and, after serum restimulation, individual cells were monitored for S phase entry by immunodetection of BrdU incorporation during the 24 h period after stimulation. Results are expressed as the percentage of GFP-positive cells incorporating BrdU. (F) Cells overexpressing both PRMT5 and cyclin E1 resume DNA synthesis. NIH 3T3 cells were co-transfected with expression plasmids encoding CD20 marker and PRMT5 together with cyclin E or an empty control vector. DNA profiles of CD20-positive cells were obtained using bivariate flow cytometry. The _y_-axis shows the increase in percentage of cells in the G0/G1 phase upon expression of the indicated proteins. The baseline represents the percentage of G0/G1 cells in mock-transfected cells.
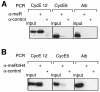
Fig. 3. ChIP analyses reveal the association of proteins methylated on arginine residues with the transcription start site region of the cyclin E1 promoter in vivo. (A) Crosslinked chromatin from G0-arrested NIH 3T3 fibroblasts was immunoprecipitated with an anti-methylarginine monoclonal antibody or a control antibody (anti-Flag epitope). One-tenth of the input chromatin and the immunoprecipitated chromatin were analysed by quantitative PCR for the presence of mouse cyclin E1 promoter fragments CE12 or CE6, or for fragments corresponding to the mouse albumin gene (Alb). (B) ChIPs performed as in (A) but using an antiserum directed against methylated histone H4 (me-R3-H4).
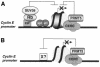
Fig. 4. Role of the type II arginine methyltransferase, PRMT5, in the repressor function of CERC. (A) Rb- and CERC-associated deacetylase, lysine and arginine methyltransferase activities maintain repression via a single nucleosome located near the transcription start site of the cyclin E1 promoter. (B) Alternatively, the target of CERC-associated methyltransferase activity is a non-nucleosomal regulatory protein located in the same region of the cyclin E1 promoter.
Similar articles
- The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene.
Cesaro E, De Cegli R, Medugno L, Florio F, Grosso M, Lupo A, Izzo P, Costanzo P. Cesaro E, et al. J Biol Chem. 2009 Nov 20;284(47):32321-30. doi: 10.1074/jbc.M109.043349. Epub 2009 Sep 9. J Biol Chem. 2009. PMID: 19741270 Free PMC article. - Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes.
Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Pal S, et al. Mol Cell Biol. 2004 Nov;24(21):9630-45. doi: 10.1128/MCB.24.21.9630-9645.2004. Mol Cell Biol. 2004. PMID: 15485929 Free PMC article. - Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression.
Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Rank G, et al. Blood. 2010 Sep 2;116(9):1585-92. doi: 10.1182/blood-2009-10-251116. Epub 2010 May 21. Blood. 2010. PMID: 20495075 Free PMC article. - Versatility of PRMT5-induced methylation in growth control and development.
Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Karkhanis V, et al. Trends Biochem Sci. 2011 Dec;36(12):633-41. doi: 10.1016/j.tibs.2011.09.001. Epub 2011 Oct 3. Trends Biochem Sci. 2011. PMID: 21975038 Free PMC article. Review. - The Structure and Function of the PRMT5:MEP50 Complex.
Antonysamy S. Antonysamy S. Subcell Biochem. 2017;83:185-194. doi: 10.1007/978-3-319-46503-6_7. Subcell Biochem. 2017. PMID: 28271477 Review.
Cited by
- Histone Arginine Methylation as a Regulator of Gene Expression in the Dehydrating African Clawed Frog (Xenopus laevis).
Rehman S, Parent M, Storey KB. Rehman S, et al. Genes (Basel). 2024 Sep 1;15(9):1156. doi: 10.3390/genes15091156. Genes (Basel). 2024. PMID: 39336747 Free PMC article. - Genetic screen identified PRMT5 as a neuroprotection target against cerebral ischemia.
Wu H, Lv P, Wang J, Bennett B, Wang J, Li P, Peng Y, Hu G, Lin J. Wu H, et al. Elife. 2024 Feb 19;12:RP89754. doi: 10.7554/eLife.89754. Elife. 2024. PMID: 38372724 Free PMC article. - The potential and challenges of targeting _MTAP_-negative cancers beyond synthetic lethality.
Bray C, Balcells C, McNeish IA, Keun HC. Bray C, et al. Front Oncol. 2023 Sep 19;13:1264785. doi: 10.3389/fonc.2023.1264785. eCollection 2023. Front Oncol. 2023. PMID: 37795443 Free PMC article. Review. - Methylation across the central dogma in health and diseases: new therapeutic strategies.
Liu R, Zhao E, Yu H, Yuan C, Abbas MN, Cui H. Liu R, et al. Signal Transduct Target Ther. 2023 Aug 25;8(1):310. doi: 10.1038/s41392-023-01528-y. Signal Transduct Target Ther. 2023. PMID: 37620312 Free PMC article. Review. - Inhibition of histone methyltransferase PRMT5 attenuates cisplatin-induced hearing loss through the PI3K/Akt-mediated mitochondrial apoptotic pathway.
Zheng Z, Nan B, Liu C, Tang D, Li W, Zhao L, Nie G, He Y. Zheng Z, et al. J Pharm Anal. 2023 Jun;13(6):590-602. doi: 10.1016/j.jpha.2023.04.014. Epub 2023 Apr 26. J Pharm Anal. 2023. PMID: 37440906 Free PMC article.
References
- Branscombe T.L., Frankel, A., Lee, J.H., Cook, J.R., Yang, Z., Pestka, S. and Clarke, S. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem., 276, 32971–32976. - PubMed
- Brehm A., Miska, E.A., McCance, D., Reid, J.L., Bannister, A. and Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. - PubMed
- Chen D., Ma, H., Hong, H., Koh, S., Huang, S.M., Schurter, B.T., Aswad, D.W. and Stallcup, M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI043369/AI/NIAID NIH HHS/United States
- R01 AI036450/AI/NIAID NIH HHS/United States
- R01-CA46465/CA/NCI NIH HHS/United States
- R01 AI36450/AI/NIAID NIH HHS/United States
- R01-AI43369/AI/NIAID NIH HHS/United States
- R01 CA046465/CA/NCI NIH HHS/United States
- 2T32AI07403/AI/NIAID NIH HHS/United States
- T32 AI007403/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases