Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15 - PubMed (original) (raw)
Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15
Isabelle C Braun et al. Mol Cell Biol. 2002 Aug.
Abstract
Metazoan NXF1/p15 heterodimers promote export of bulk mRNA through nuclear pore complexes (NPC). NXF1 interacts with the NPC via two distinct structural domains, the UBA-like domain and the NTF2-like scaffold, which results from the heterodimerization of the NTF2-like domain of NXF1 with p15. Both domains feature a single nucleoporin-binding site, and they act synergistically to promote NPC translocation. Whether the NTF2-like scaffold (and thereby p15) contributes only to NXF1/NPC association or is also required for other functions, e.g., to impart directionality to the export process by regulating NXF1/NPC or NXF1/cargo interactions, remains unresolved. Here we show that a minimum of two nucleoporin-binding sites is required for NXF1-mediated export of cellular mRNA. These binding sites can be provided by an NTF2-like scaffold followed by a UBA-like domain (as in the wild-type protein) or by two NTF2-like scaffolds or two UBA-like domains in tandem. In the latter case, the export activity of NXF1 is independent of p15. Thus, as for the UBA-like domain, the function of the NTF2-like scaffold is confined to nucleoporin binding. More importantly, two copies of either of these domains are sufficient to promote directional transport of mRNA cargoes across the NPC.
Figures
FIG. 1.
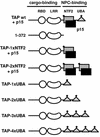
Structural and functional domains of human TAP bound to p15. The cargo-binding domain of TAP (residues 1 to 372) consists of an RNP-type RNA-binding domain (RBD, residues 119 to 198), a leucine-rich repeat domain (LRR, residues 203 to 362), and a less well conserved region N-terminal to the RBD for which no structural information is available (residues 1 to 119). The NPC-binding domain of TAP consists of the NTF2-like domain (grey square, residues 371 to 551) bound to p15 (black square) and a UBA-like domain (triangle, residues 563 to 619). Each of these domains features a nucleoporin-binding site (solid diamond).
FIG. 2.
Two nucleoporin-binding domains are required for TAP-mediated translocation of RNP cargoes across the NPC. (A) Schematic representation of the cat reporter gene encoded by plasmid pCMV128 (16). SD and SA indicate the splice donor and splice acceptor sites of the intron, respectively. (B) Human 293 cells were transfected with a mixture of plasmids encoding β-Gal (pCH110), CAT (pCMV128), and either GFP alone or GFP fused N-terminally to TAP or a TAP mutant, as indicated on the left. When indicated, a pGFP-N3 derivative encoding zzp15 was cotransfected (+p15, black bars). Cells were collected 48 h after transfection, and β-Gal and CAT activities were determined. CAT activities were normalized to the activities of the β-Gal internal control. The stimulation of cat gene expression by TAP mutants measured in three independent experiments is expressed as a percentage of the activity of wild-type TAP. The values are means ± standard deviations. (C) Protein expression levels were analyzed by Western blot with anti-GFP antibodies. The amount of cell extract loaded per lane was corrected for minor differences in transfection efficiencies, as revealed by the β-Gal internal control. The positions of TAP derivatives fused to GFP or of zzp15 are shown on the right. (D) Human 293 cells were transfected with the reporter plasmids pCMV128-RRE and pCH110 and plasmids encoding RevM10 fusions of TAP, TAP-1xUBA, or TAP-2xUBA. When indicated, a plasmid encoding zzp15 (+p15) was cotransfected (black bar). Thereporter plasmid pCMV128-RRE is identical to pCMV128 (panel A) but carries the HIV RRE inserted in the intron, downstream of the cat gene. The stimulation of cat gene expression by TAP mutants measured in three independent experiments is expressed as a percentage of the activity of RevM10-TAP coexpressed with p15. The values are means ± standard deviations. (E) Protein expression levels were analyzed by Western blot with anti-HA antibodies. The amount of cell extract loaded per lane was normalized to the activity of the β-Gal internal control. The positions of TAP derivatives fused to RevM10 are shown on the right. (F) RNase protection analysis was performed with cytoplasmic or total RNA fractions isolated from 293 cells cotransfected with pCMV128 and plasmid pCH110 encoding β-Gal. In lanes 3, 5, 8, and 10, plasmids encoding p15 and RevM10 or GFP fusions of TAP were cotransfected. As controls, the reporter plasmids were cotransfected with empty vectors expressing RevM10 or GFP alone together with the plasmid expressing p15 (lanes 2, 4, 7, and 9).
FIG. 3.
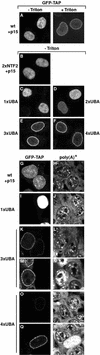
TAP-3xUBA and TAP-4xUBA localize to the nuclear rim. (A) HeLa cells were transfected with a plasmid expressing GFP-TAP. When cells were directly fixed in formaldehyde, TAP was predominantly detected throughout the nucleoplasm, excluding nucleoli (−Triton). When cells were extracted with Triton X-100 prior to fixation, a punctate labeling pattern was visible at the nuclear periphery (+Triton). (B to F) HeLa cells were transfected with plasmids expressing GFP fusions of TAP derivatives as indicated. Approximately 20 h after transfection, cells were fixed in formaldehyde and directly observed with a confocal microscope. TAP-2xNTF2, TAP-1xUBA, and TAP-2xUBA were detected throughout the nucleoplasm (B, C, and D). An additional punctate labeling pattern was visible at the nuclear rim for TAP-2xUBA. TAP-3xUBA and TAP-4xUBA localized predominantly at the nuclear rim (E, F). (G to R) Human 293 cells were transfected with plasmids encoding GFP fusions of TAP derivatives as indicated. Approximately 20 h after transfection, bulk poly(A)+ RNA was detected by FISH with an indocarbocyanine-labeled oligo(dT) probe. All images were taken with the same settings so the GFP signals could be compared directly. In panels K, M, O, and Q, examples of cells expressing TAP-3xUBA or TAP-4xUBA at different levels are shown. In about 10 to 20% of transfected cells, TAP-3xUBA and TAP-4xUBA were expressed at higher levels (M and Q), resulting in a strong accumulation of the poly(A)+ signal within the nucleus (N and R). wt, wild type.
FIG. 4.
TAP-2xUBA and TAP-3xUBA stimulate mRNA export in Xenopus oocytes. (A and B) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) andcytoplasmic (C) fractions were collected 120 min after injection or immediately after injection (t0; lanes 1 to 3 in panel B). RNA samples were analyzed on 8% acrylamide-7 M urea denaturing gels. One oocyte equivalent of RNA, from a pool of 10 oocytes, was loaded per lane. The numbers above the lanes in panel A indicate the concentration (micromolar) of recombinant proteins in the injected samples. The numbers in boxes below the panels represent the percentage of a given RNA in the cytoplasm 120 min after injection in the presence of the proteins indicated above the lanes. Because the nuclear localization of U6Δss RNA and the export of tRNA (>94%) were not affected by any of the injected proteins, the respective values are not included. On the right of panel A, the effects of increasing concentrations of TAP-2xUBA or TAP-3xUBA on the export of U5ΔSm RNA and Ftz and Ad mRNAs are compared to those of TAP/p15 heterodimers. For the purpose of comparison, all values were normalized to the export observed in control oocytes. (C) TAP derivatives were expressed in E. coli as GST fusions, purified on glutathione-agarose beads, and visualized by Coomassie staining following SDS-polyacrylamide gel electrophoresis. (D) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) and cytoplasmic (C) fractions were collected 120 min after injection or immediately after injection (t0; lanes 1 to 3) and analyzed as in panels A and B. DHFR, β-globin, and Ftz mRNAs exhibited similar behavior, so only the quantitation for DHFR mRNA is shown. The nuclear localization of U6Δss RNA was not affected by any of the injected proteins.
FIG. 5.
TAP derivatives having a single nucleoporin-binding domain translocate specific CTE RNA cargoes across the NPC. (A and B) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) and cytoplasmic (C) fractions were collected immediately after injection (t0; lanes 1 to 3) or 90 min after injection in panel A and analyzed as described in the legend to Fig. 4. In panel B, samples were collected 120 min after injection and analyzed in 10% acrylamide-7 M urea denaturing gels. The concentration of recombinant proteins in the injected samples was 3 μM. The numbers below the panels represent the percentage of a given RNA in the cytoplasm in the presence of the proteins indicated above the lanes. The nuclear localization of U6Δss RNA and the export of tRNA (>94%) were not affected by any of the injected proteins, and these values are not included. The mature products and intermediates of the splicing reaction are indicated diagrammatically on the left of panel B. The solid triangle represents the M38 CTE. wt, wild type.
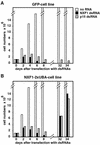
FIG. 6.
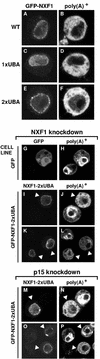
D. melanogaster NXF1-2xUBA partially rescues export in cells depleted of NXF1. (A to F) SL2 cell lines constitutively expressing GFP fusions with wild-type NXF1 (wt), NXF1-1xUBA, or NXF1-2xUBA were fixed in formaldehyde. Bulk poly(A)+ RNA was detected by FISH (B, D, and F). (G to P) SL2 cells stably transfected with plasmids expressing GFP or GFP-NXF1-2xUBA were transfected with a dsRNA corresponding to the NTF2-like domain of D. melanogaster NXF1 (G to L) or to the coding region of D. melanogaster p15(M to P). Five days after transfection, poly(A)+ RNA was detected by FISH with an indocarbocyanine-labeled oligo(dT) probe. In the GFP-expressing cell line, depletion of NXF1 causes nuclear accumulation of poly(A)+ RNA regardless of whether GFP is expressed (G and H). In the GFP-NXF1-2xUBA cell line, a strong nuclear accumulation of poly(A)+ RNA was observed in cells in which the protein was not detected, but when D. melanogaster NXF1-2xUBA was expressed at detectable levels, mRNA export was restored (I to L, arrowheads). This restoration of export was also observed when p15 was depleted (M to P).
FIG. 7.
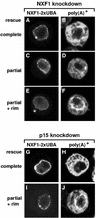
Patterns of poly(A)+ RNA distribution in cells depleted of endogenous NXF1 or p15 but expressing GFP-NXF1-2xUBA. (A to J) Restoration of mRNA export was almost complete (B and H) or partial, with an equal distribution of the oligo(dT) signal between the nucleus and the cytoplasm (D). In panels F and J, cells in which the poly(A)+ RNA accumulated at the nuclear rim are shown.
FIG. 8.
NXF1-2xUBA sustains growth of cells depleted of endogenous NXF1 or p15. (A and B) SL2 cells stably transfected with plasmids expressing GFP or GFP-NXF1-2xUBA were transfected with NXF1 or p15 dsRNAs as indicated. Cell numbers were determined every second day for 8 days and every week for 6 weeks. In the GFP cell line, no cells survived NXF1 depletion. The surviving cells detected 31 days following p15 depletion died progressively after a few days. Cells expressing NXF1-2xUBA survived NXF1 or p15 depletion and started to grow exponentially 1 month after transfection of the dsRNAs.
Similar articles
- RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export.
Forler D, Rabut G, Ciccarelli FD, Herold A, Köcher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E. Forler D, et al. Mol Cell Biol. 2004 Feb;24(3):1155-67. doi: 10.1128/MCB.24.3.1155-1167.2004. Mol Cell Biol. 2004. PMID: 14729961 Free PMC article. - Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Fribourg S, Braun IC, Izaurralde E, Conti E. Fribourg S, et al. Mol Cell. 2001 Sep;8(3):645-56. doi: 10.1016/s1097-2765(01)00348-3. Mol Cell. 2001. PMID: 11583626 - Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export.
Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Braun IC, et al. J Biol Chem. 2001 Jun 8;276(23):20536-43. doi: 10.1074/jbc.M100400200. Epub 2001 Mar 19. J Biol Chem. 2001. PMID: 11259411 - A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm.
Izaurralde E. Izaurralde E. Eur J Cell Biol. 2002 Nov;81(11):577-84. doi: 10.1078/0171-9335-00273. Eur J Cell Biol. 2002. PMID: 12498157 Review. - Nucleocytoplasmic transport enters the atomic age.
Conti E, Izaurralde E. Conti E, et al. Curr Opin Cell Biol. 2001 Jun;13(3):310-9. doi: 10.1016/s0955-0674(00)00213-1. Curr Opin Cell Biol. 2001. PMID: 11343901 Review.
Cited by
- Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15.
Fribourg S, Conti E. Fribourg S, et al. EMBO Rep. 2003 Jul;4(7):699-703. doi: 10.1038/sj.embor.embor883. EMBO Rep. 2003. PMID: 12835756 Free PMC article. - Nuclear export of mRNA molecules studied by SPEED microscopy.
Li Y, Junod SL, Ruba A, Kelich JM, Yang W. Li Y, et al. Methods. 2019 Jan 15;153:46-62. doi: 10.1016/j.ymeth.2018.08.005. Epub 2018 Aug 18. Methods. 2019. PMID: 30125665 Free PMC article. Review. - RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export.
Forler D, Rabut G, Ciccarelli FD, Herold A, Köcher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E. Forler D, et al. Mol Cell Biol. 2004 Feb;24(3):1155-67. doi: 10.1128/MCB.24.3.1155-1167.2004. Mol Cell Biol. 2004. PMID: 14729961 Free PMC article. - Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP.
Erkmann JA, Sànchez R, Treichel N, Marzluff WF, Kutay U. Erkmann JA, et al. RNA. 2005 Jan;11(1):45-58. doi: 10.1261/rna.7189205. RNA. 2005. PMID: 15611298 Free PMC article. - Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport.
Terry LJ, Wente SR. Terry LJ, et al. Eukaryot Cell. 2009 Dec;8(12):1814-27. doi: 10.1128/EC.00225-09. Epub 2009 Oct 2. Eukaryot Cell. 2009. PMID: 19801417 Free PMC article. Review.
References
- Bachi, A., I. C. Braun, J. P. Rodrigues, N. Panté, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Görlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. - PMC - PubMed
- Bayliss, R., K. Ribbeck, D. Akin, H. M. Kent, C. M. Feldherr, D. Görlich, and M. Stewart. 1999. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293:579-593. - PubMed
- Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin b in nuclear trafficking. Cell 102:99-108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous