TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4 - PubMed (original) (raw)
TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4
Sohail Malik et al. Mol Cell Biol. 2002 Aug.
Abstract
The orphan nuclear receptor hepatocyte nuclear factor 4 (HNF-4) regulates the expression of many liver-specific genes both during development and in the adult animal. Towards understanding the molecular mechanisms by which HNF-4 functions, we have established in vitro transcription systems that faithfully recapitulate HNF-4 activity. Here we have focused on the coactivator requirements for HNF-4, especially for the multicomponent TRAP/SMCC/Mediator complex that has emerged as the central regulatory module of the transcription apparatus. Using a system that has been reconstituted from purified transcription factors, as well as one consisting of unfractionated nuclear extract from which TRAP/SMCC/Mediator has been depleted by specific antibodies, we demonstrate a strong dependence of HNF-4 function on this coactivator. Importantly, we further show a TRAP/SMCC/Mediator-dependence for HNF-4 transcriptional activation from chromatin templates. The latter involves cooperation with the histone acetyltransferase-containing coactivator p300, in accord with a synergistic mode of action of the two divergent coactivators. We also show that HNF-4 and TRAP/SMCC/Mediator can interact physically. This interaction likely involves primary HNF-4 activation function 2 (AF-2)-dependent interactions with the TRAP220 subunit of TRAP/SMCC/Mediator and secondary (AF-2-independent) interactions with TRAP170/RGR1. Finally, recruitment experiments using immobilized templates strongly suggest that the functional consequences of the physical interaction probably are manifested at a postrecruitment step in the activation pathway.
Figures
FIG. 1.
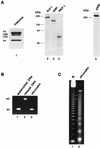
TRAP/SMCC/Mediator-dependent transcriptional activation of DNA templates by HNF-4. (A) TRAP/SMCC/Mediator-dependent function of HNF-4 in a transcription system reconstituted from purified factors. In vitro transcription reaction mixtures were reconstituted with 50 ng of TFIIA, 10 ng of TFIIB, 10 ng of TFIIEα, 5 ng of TFIIEβ, 25 ng of TFIIF, 20 ng of TFIIH, 50 ng of Pol II, 150 ng of PC4, and an amount of affinity-purified TFIID containing 5 μg of TBP. GAL4-AH (25 ng) was added to reaction mixtures in lanes 3 and 4; HNF-4 (50 ng) was added to reaction mixtures in lanes 5 and 6. Purified TRAP/SMCC/Mediator (f: NUT2) was included in reaction mixtures in lanes 2, 4, and 6. Reaction mixtures also contained 50 ng each of the templates pG5HML and pA4xMLΔ53. (B) Depletion of TRAP/SMCC/Mediator from HeLa nuclear extract. HeLa nuclear extract was incubated with control beads (lane 1) or with beads containing cross-linked anti-NUT2 antibodies (lane 2). Unbound extract was immunoblotted with indicated antibodies. (C) TRAP/SMCC/Mediator-dependent function of HNF-4 in nuclear extract. In vitro transcription reaction mixtures contained a control nuclear extract (lanes 1 to 4) or TRAP/SMCC/Mediator-depleted extract (lanes 5 to 8). HNF-4 was added to reaction mixtures in lanes 2, 4, 6, and 8. Purified TRAP/SMCC/Mediator was added to reaction mixtures in lanes 3, 4, 7, and 8.
FIG. 2.
Reconstitution of chromatin on templates containing HNF-4 cognate sites. (A) Purified preparations of the indicated proteins used for reconstituting chromatin on plasmid pA4xMLΔ53 were analyzed by SDS-PAGE and stained with Coomassie brilliant blue (lanes 1 and 5) or silver (lanes 2 to 4). Lane 1, HeLa cell histones, 1.5 μg; lane 2, baculovirus-expressed Acf-1, 200 ng; lane 3, baculovirus-expressed ISWI, 200 ng; lane 4, bacterially expressed NAP-1, 200 ng; lane 5, baculovirus-expressed p300, 1 μg. (B) DNA supercoiling assay for chromatin assembly on plasmid pA4xMLΔ53. Plasmid DNA, treated as described, was electrophoresed on agarose gels and stained with ethidium bromide. Lane 1, purified plasmid prior to assembly; lane 2, topoisomerase I-relaxed plasmid; lane 3, plasmid after chromatin assembly (and deproteinization). Supercoiled (sc) and relaxed (rel) DNAs are marked. (C) MNase digestion assay for chromatin assembly. After assembly into chromatin, plasmid pA4xMLΔ53 was digested with MNase, deproteinized, and analyzed by agarose gel electrophoresis. A 123-bp ladder (M) was used as a size marker.
FIG. 3.
HNF-4 function on chromatin templates is dependent on TRAP/SMCC/Mediator and is stimulated by p300. In vitro transcription reactions contained chromatinized plasmid pA4xMLΔ53 (40 ng) and plasmid pG5HML (50 ng) as naked DNA (lanes 3 to 14). The templates were preincubated with HNF-4 (50 ng, lanes 2, 4, 6, 8, 10, 12, and 14) and p300 (200 ng, lanes 5, 6, 9, 10, 13, and 14) for 25 min. Nuclear extract (control, lanes 1 to 6; or TRAP/SMCC/Mediator-depleted [ΔMED], lanes 7 to 14) was then added, and incubation continued for 20 min. To reaction mixtures in lanes 12 and 14, purified TRAP/SMCC/Mediator was also added at this time. Labeled nucleotide triphosphate mix was then added, and after 30 min, the samples were processed for analysis by electrophoresis. Reaction mixtures in lanes 1 and 2 contained naked pA4xMLΔ53 as a control.
FIG. 4.
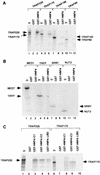
Direct physical interaction between HNF-4 and TRAP/SMCC/Mediator. (A) Schematic representation of full-length HNF-4 and various derivatives used in GST interaction assays (see text for details). Relevant features only are shown. DBD, DNA binding domain; P-rich, proline-rich; AF-1 and AF-2, activation functions 1 and 2, respectively. (B) Purified TRAP/SMCC/Mediator (10% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). After being washed to remove unbound material, the resins were eluted with Sarkosyl and analyzed by immunoblotting following SDS-PAGE.
FIG. 5.
Identification of TRAP/SMCC/Mediator subunits that interact with HNF-4. (A) Selected subunits were expressed as 35S-labeled proteins in a rabbit reticulocyte coupled transcription and translation system, incubated with GST alone or with GST-HNF-4ΔC1 (as indicated) and processed as described in the legend to Fig. 4B. Lanes: 1 to 3, TRAP220; 4 to 6, TRAP170/RGR1; 7 to 9, TRAP100; 10 to 12, TRAP80. Inputs (in) representing 20% of the amount used in the binding reaction are also shown. (B) Additional (low-molecular-weight) selected subunits were expressed as 35S-labeled proteins in a rabbit reticulocyte coupled transcription and translation system, incubated with GST alone or with GST-HNF-4ΔC1 (as indicated), and processed as described in the legend to Fig. 4B. Lanes: 1 to 3, MED7; 4 to 6, TRFP; 7 to 9, SRB7; 10 to 12, NUT2. (C) HNF-4 AF-2-dependent and AF-2-independent interactions with TRAP/SMCC/Mediator subunits. 35S-labeled TRAP220 (lanes 1 to 5) and TRAP170/RGR1 (lanes 6 to 10) were incubated with GST alone (lanes 2 and 7), GST-HNF-4ΔC1 (lanes 3 and 8), GST-HNF-4ΔC2 (lanes 4 and 9), or GST-HNF-4-LBD (lanes 5 and 10) and processed as described in the legend to Fig. 4B.
FIG. 6.
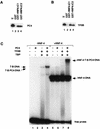
PC4-dependent stabilization of PIC. (A) Purified recombinant PC4 (25% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). Samples were processed as described in the legend to Fig. 4B. Following SDS-PAGE, PC4 was detected by immunoblotting. (B) Purified recombinant TFIIB (25% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). Samples were processed as described in the legend to Fig. 4B. Following SDS-PAGE, TFIIB was detected by immunoblotting. (C) For EMSA, an end-labeled probe consisting of site A and the apolipoprotein AI core promoter (29) was incubated with the indicated combinations of factors: PC4 (75 ng, lanes 2, 4, 6, and 8); HNF-4 (50 ng, lanes 5 to 8); TBP (T, 0.5 ng, lanes 3, 4, 7, and 8); TFIIB (B, 5 ng, lanes 4 and 8). Although the description of the PC4-dependent EMSA complexes implies that they contain PC4, it is currently not possible to rigorously demonstrate this fact. In a previous analysis (27), antibodies directed against PC4 failed to supershift the complex designated TBP-TFIIB-PC4 either because the PC4 epitopes are not accessible (by virtue of multiple contacts) or because PC4 association with the complex fails to survive electrophoresis. Nonetheless, we can show that PC4 is present in analogous complexes formed on immobilized templates (Fig. 7).
FIG. 7.
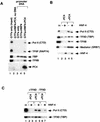
Recruitment of the transcription machinery into the PIC. (A) Complexes were assembled on M280-streptavidin Dynabeads carrying a biotinylated DNA fragment from the plasmid pA4xMLΔ53, which contained four copies of the HNF-4 cognate site A and the adenovirus major late core promoter. After PIC formation, beads were washed and bound complexes were analyzed for their factor content by immunoblotting. PICs were formed with GTFs only (TBP, TFIIB, TFIIE, TFIIF, TFIIH, Pol II (lane 3); with GTFs and HNF-4 (lane 4); or with GTFs, HNF-4, and PC4 (lanes 2 and 5). Lane 2, control in which the indicated factors were incubated with beads only (no template DNA); lane 1, GTFs only (input). (B) Immobilized-template recruitment assays are as for panel A, except that all reaction mixtures contained TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Pol II, and TRAP/SMCC/Mediator, as shown in input lane 1. HNF-4 (lanes 3 and 5) and PC4 (lanes 4 and 5) were additionally added as indicated. (C) Immobilized-template recruitment assays are as for panel B, except that reaction mixtures in lanes 6 and 7 did not contain TFIID. HNF-4 (lanes 3, 5, and 7) and PC4 (lanes 4 to 7) were additionally added as indicated.
Similar articles
- Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins.
Soutoglou E, Papafotiou G, Katrakili N, Talianidis I. Soutoglou E, et al. J Biol Chem. 2000 Apr 28;275(17):12515-20. doi: 10.1074/jbc.275.17.12515. J Biol Chem. 2000. PMID: 10777539 - The TRAP/SMCC/Mediator complex and thyroid hormone receptor function.
Ito M, Roeder RG. Ito M, et al. Trends Endocrinol Metab. 2001 Apr;12(3):127-34. doi: 10.1016/s1043-2760(00)00355-6. Trends Endocrinol Metab. 2001. PMID: 11306338 Review. - Erythropoietin gene regulation depends on heme-dependent oxygen sensing and assembly of interacting transcription factors.
Huang LE, Ho V, Arany Z, Krainc D, Galson D, Tendler D, Livingston DM, Bunn HF. Huang LE, et al. Kidney Int. 1997 Feb;51(2):548-52. doi: 10.1038/ki.1997.76. Kidney Int. 1997. PMID: 9027736 Review.
Cited by
- Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator.
Malik S, Baek HJ, Wu W, Roeder RG. Malik S, et al. Mol Cell Biol. 2005 Mar;25(6):2117-29. doi: 10.1128/MCB.25.6.2117-2129.2005. Mol Cell Biol. 2005. PMID: 15743810 Free PMC article. - Proteomics reveals a physical and functional link between hepatocyte nuclear factor 4alpha and transcription factor IID.
Takahashi H, Martin-Brown S, Washburn MP, Florens L, Conaway JW, Conaway RC. Takahashi H, et al. J Biol Chem. 2009 Nov 20;284(47):32405-12. doi: 10.1074/jbc.M109.017954. Epub 2009 Oct 5. J Biol Chem. 2009. PMID: 19805548 Free PMC article. - Gene-specific transcription activation via long-range allosteric shape-shifting.
Tsai CJ, Nussinov R. Tsai CJ, et al. Biochem J. 2011 Oct 1;439(1):15-25. doi: 10.1042/BJ20110972. Biochem J. 2011. PMID: 21916844 Free PMC article. Review. - TAF4, a subunit of transcription factor II D, directs promoter occupancy of nuclear receptor HNF4A during post-natal hepatocyte differentiation.
Alpern D, Langer D, Ballester B, Le Gras S, Romier C, Mengus G, Davidson I. Alpern D, et al. Elife. 2014 Sep 10;3:e03613. doi: 10.7554/eLife.03613. Elife. 2014. PMID: 25209997 Free PMC article. - Transcription regulation by the Mediator complex.
Soutourina J. Soutourina J. Nat Rev Mol Cell Biol. 2018 Apr;19(4):262-274. doi: 10.1038/nrm.2017.115. Epub 2017 Dec 6. Nat Rev Mol Cell Biol. 2018. PMID: 29209056 Review.
References
- Arias, J. A., and W. S. Dynan. 1989. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J. Biol. Chem. 264:3223-3229. - PubMed
- Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. - PubMed
- Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. - PubMed
- Chen, W. S., K. Manova, D. C. Weinstein, S. A. Duncan, A. S. Plump, V. R. Prezioso, R. F. Bachvarova, and J. E. Darnell, Jr. 1994. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8:2466-2477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous