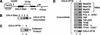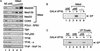TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA - PubMed (original) (raw)
TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA
Kristina M Johnson et al. Genes Dev. 2002.
Abstract
Activator-mediated transcription complex assembly on templates lacking chromatin requires the interaction of activators with two major coactivator complexes: TFIID and mediator. Here we employed immobilized template assays to correlate transcriptional activation with mediator and TFIID recruitment. In reactions reconstituted with purified proteins, we found that activator, TFIID, and mediator engage in reciprocal cooperative interactions to form a complex on promoter DNA. Preassembly of the coactivator complex accelerates the rate of transcription in a cell-free system depleted of TFIID and mediator. Our data argue that this coactivator complex is an intermediate in the assembly of an active transcription complex. Furthermore, the reciprocity of the interactions demonstrates that the complex could in principle be nucleated with either TFIID or mediator, implying that alternative pathways could be utilized to generate diversity in the way activators function in vivo.
Figures
Figure 1
Transcriptional activation by GAL4-VP16 correlates with recruitment of the general machinery. (A) Schematic of the 650-bp immobilized template bearing five GAL4 DNA binding sites 23 bp upstream of the adenovirus E4 TATA box. The _Hin_dIII site is used to cleave the DNA from the Dynabeads™ for quantitation, by agarose gel electrophoresis. (B) The amount of GAL4-VP16 protein bound to the immobilized templates is proportional to the number of GAL4 DNA binding sites. Two hundred ng purified, recombinant GAL4-VP16 was added to 25 fmol immobilized template bearing 0, 1, 2, or 5 GAL4 DNA binding sites. The bound protein was fractionated by SDS-PAGE. An immunoblot of GAL4-VP16 is shown. (C) GAL4-VP16 stimulates activated levels of transcription from the immobilized templates. One hundred-twenty micrograms of HeLa nuclear extract was added in batch with 5 ng GAL4-VP16 and 10 fmol immobilized DNA template. After 1 h at 30°C, the transcription products were analyzed by primer extension analysis (arrows denote extension product) and resolved on a 10% polyacrylamide/urea gel. A phosphorimager scan of the gel is shown. (D) GAL4-VP16 stimulates recruitment of general transcription factors and the human mediator coactivator complex to immobilized templates. One hundred-twenty micrograms of HeLa nuclear extract was incubated with 10 fmol immobilized DNA template in the absence (lane 1) or presence (lane 2) of 5 ng GAL4-VP16. After 1 h at room temperature, the beads were washed and bound protein was fractionated on 4%–15% SDS polyacrylamide gels. Individual factors were assayed by immunoblot. Autoradiographs are shown from several experiments measuring different sets of factors.
Figure 2
Composition and activity of purified human mediator (hMed) and mediator-depleted nuclear extract (ΔNE). (A) HeLa nuclear extracts were fractionated on a GST-TRLBD affinity matrix. The starting (lane 1) and depleted extract (lane 2) along with increasing amounts of mediator fraction (lanes 3_–_5) were fractionated on 4%–15% SDS polyacrylamide gels and analyzed by immunoblot with antibodies against representative subunits of hMed and GTFs. Autoradiographs of the blots are shown. Note that hMed is depleted from the ΔNE. Med220 is depleted greater than 30-fold. Also, general transcription factors, including TBP and TAFs of TFIID, are present in the ΔNE fraction but are not present in the purified hMed fraction. (B) Purified hMed complements ΔNE for activated transcription. Twenty nanograms of pG5E4T template (lanes 1_–_6) was incubated with 35 μg ΔNE (lanes 1_–_4) and (lanes 3,4) or 1 unit purified hMed (see Materials and Methods) (lanes 5,6) in the absence (lanes 1,3,5) or presence (lanes 2,4,6) of 5 ng GAL4-VP16. Transcription was measured by primer extension (arrow). Note that hMed alone cannot support transcription. GAL4-VP16-induced transcription is detected only when purified hMed is supplemented with ΔNE (lane 4). (C) The VP eluate complements ΔNE. pG5E4T (lanes 1_–_8) was incubated with 35 μg HeLa NE (lanes 1,2), 35 μg ΔNE (lanes 3_–_6) and/or 2μg VP eluate (lanes 4_–_8) in the presence (+) or absence (−) of GAL4-VP16. Note that neither VP eluate nor ΔNE alone support transcription but the combination is active.
Figure 3
hMed is recruited by GAL4-VP16 on immobilized templates. (A) GAL4-VP16 stimulates recruitment of purified hMed to immobilized DNA templates. One unit purified hMed was incubated with 5 ng purified, recombinant GAL4-VP16 and 10 fmol immobilized G5E4T DNA. Bound protein is detected by immunoblot. (B) Purified hMed is required for activation on immobilized templates. Twenty nanograms of pG5E4T template (lanes 1_–_4) was incubated with 35 μg ΔNE (lanes 1_–_4) and (lanes 3,4) 1 unit purified hMed in the absence (lanes 1,3) or presence (lanes 2,4) of 5 ng GAL4-VP16. A phosphorimager scan of the gel is shown.
Figure 4
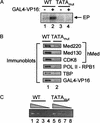
Cooperative assembly of transcription complexes requires a TATA box. (A) In vitro transcription of G5E4T requires a TATA box. HeLa NE (lanes 1_–_4) was incubated with 40 fmol WT G5E4T immobilized template (lanes 1,2) or 40 fmol TATAmut G5E4T immobilized template (lanes 3,4). No transcription signal is detectable from the TATAmut template. (B) Recruitment of hMed by GAL4-VP16 to G5E4T immobilized templates is dependent on a TATA box. HeLa nuclear extract was added with 5 ng GAL4-VP16 (lanes 1,2) to 5 fmol immobilized WT (lane 1) or TATAmut (lane 2) template. The presence of hMed subunits and GTFs was detected by immunoblot analysis. (C) Quantitation of immobilized templates. _Hin_dIII digests of WT or TATAmut G5E4T immobilized templates were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide. Four steps of twofold dilutions of the WT and TATAmut digests are shown adjacent to each other (WT, lanes 1_–_4; TATAmut, lanes 5_–_8).
Figure 5
Human mediator and TFIID bind cooperatively to promoter DNA. (A) Purified hMed and TFIID are active in standard in vitro transcription reactions. Mediator-depleted, TFIID-depleted nuclear extract (ΔΔNE) was generated by heat-treatment of ΔNE. The transcription reactions contained 20 ng pG5E4T with (+) or without (−) 5 ng GAL4-VP16 and 48 μg HeLa NE (lanes 1,2), 36 μg ΔNE (lanes 3_–_8), or 35 μg ΔΔNE (lanes 11_–_18) in the presence of 120 ng TFIID (lanes 7,8,13,14,17,18) and 1 unit hMed (lanes 5_–_10,15_–_18). Transcription was analyzed by primer extension (arrow). An autoradiograph of the gel is shown. (B) Saturating levels of purified hMed recruit TFIID to immobilized templates. Twenty-five fmol G5E4T immobilized template (every lane) was prebound with 200 ng GAL4-VP16 (+) in every other lane. Subsequently, 40 ng TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6. In lanes 3_–_6, 0.8 units hMed were added. After washing, bound proteins were detected by immunoblot analysis. The approximate stoichiometry of GAL4-VP16, TFIID, and hMed recruitment is 10 (5 GAL4-VP16 dimers):1:1, respectively (see Materials and Methods). (C) Saturating levels of TFIID stimulate hMed recruitment to immobilized templates. Twenty-five fmol G5E4T templates were prebound with 200 ng GAL4-VP16 in every other lane. One hundred-twenty nanograms of TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6; 0.04 units hMed were added to lanes 3_–_6. Protein bound to the immobilized templates was detected by immunoblot. (D) hMed (from the VP eluate) and purified TFIID bind cooperatively to immobilized G5E4T templates. Immobilized template recruitment assay: 25 fmol of immobilized G5E4T was preincubated for 20 min at room temperature in the presence (+) and absence (−) of 200 ng GAL4-VP16 with indicated amounts of VP eluate, and/or TFIID (40 ng) and TFIIA (12 ng). After 30 min, unbound protein was removed by washing and the bound protein was resolved on 4%–15% SDS-PAGE. Immunoblots of bound protein are shown. Med130 is used to measure hMed binding; TBP (HA) is representative of TFIID binding. Immunoblots with other subunits of these protein complexes demonstrate identical results.
Figure 5
Human mediator and TFIID bind cooperatively to promoter DNA. (A) Purified hMed and TFIID are active in standard in vitro transcription reactions. Mediator-depleted, TFIID-depleted nuclear extract (ΔΔNE) was generated by heat-treatment of ΔNE. The transcription reactions contained 20 ng pG5E4T with (+) or without (−) 5 ng GAL4-VP16 and 48 μg HeLa NE (lanes 1,2), 36 μg ΔNE (lanes 3_–_8), or 35 μg ΔΔNE (lanes 11_–_18) in the presence of 120 ng TFIID (lanes 7,8,13,14,17,18) and 1 unit hMed (lanes 5_–_10,15_–_18). Transcription was analyzed by primer extension (arrow). An autoradiograph of the gel is shown. (B) Saturating levels of purified hMed recruit TFIID to immobilized templates. Twenty-five fmol G5E4T immobilized template (every lane) was prebound with 200 ng GAL4-VP16 (+) in every other lane. Subsequently, 40 ng TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6. In lanes 3_–_6, 0.8 units hMed were added. After washing, bound proteins were detected by immunoblot analysis. The approximate stoichiometry of GAL4-VP16, TFIID, and hMed recruitment is 10 (5 GAL4-VP16 dimers):1:1, respectively (see Materials and Methods). (C) Saturating levels of TFIID stimulate hMed recruitment to immobilized templates. Twenty-five fmol G5E4T templates were prebound with 200 ng GAL4-VP16 in every other lane. One hundred-twenty nanograms of TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6; 0.04 units hMed were added to lanes 3_–_6. Protein bound to the immobilized templates was detected by immunoblot. (D) hMed (from the VP eluate) and purified TFIID bind cooperatively to immobilized G5E4T templates. Immobilized template recruitment assay: 25 fmol of immobilized G5E4T was preincubated for 20 min at room temperature in the presence (+) and absence (−) of 200 ng GAL4-VP16 with indicated amounts of VP eluate, and/or TFIID (40 ng) and TFIIA (12 ng). After 30 min, unbound protein was removed by washing and the bound protein was resolved on 4%–15% SDS-PAGE. Immunoblots of bound protein are shown. Med130 is used to measure hMed binding; TBP (HA) is representative of TFIID binding. Immunoblots with other subunits of these protein complexes demonstrate identical results.
Figure 5
Human mediator and TFIID bind cooperatively to promoter DNA. (A) Purified hMed and TFIID are active in standard in vitro transcription reactions. Mediator-depleted, TFIID-depleted nuclear extract (ΔΔNE) was generated by heat-treatment of ΔNE. The transcription reactions contained 20 ng pG5E4T with (+) or without (−) 5 ng GAL4-VP16 and 48 μg HeLa NE (lanes 1,2), 36 μg ΔNE (lanes 3_–_8), or 35 μg ΔΔNE (lanes 11_–_18) in the presence of 120 ng TFIID (lanes 7,8,13,14,17,18) and 1 unit hMed (lanes 5_–_10,15_–_18). Transcription was analyzed by primer extension (arrow). An autoradiograph of the gel is shown. (B) Saturating levels of purified hMed recruit TFIID to immobilized templates. Twenty-five fmol G5E4T immobilized template (every lane) was prebound with 200 ng GAL4-VP16 (+) in every other lane. Subsequently, 40 ng TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6. In lanes 3_–_6, 0.8 units hMed were added. After washing, bound proteins were detected by immunoblot analysis. The approximate stoichiometry of GAL4-VP16, TFIID, and hMed recruitment is 10 (5 GAL4-VP16 dimers):1:1, respectively (see Materials and Methods). (C) Saturating levels of TFIID stimulate hMed recruitment to immobilized templates. Twenty-five fmol G5E4T templates were prebound with 200 ng GAL4-VP16 in every other lane. One hundred-twenty nanograms of TFIID and 12 ng TFIIA were added to lanes 1, 2, 5, and 6; 0.04 units hMed were added to lanes 3_–_6. Protein bound to the immobilized templates was detected by immunoblot. (D) hMed (from the VP eluate) and purified TFIID bind cooperatively to immobilized G5E4T templates. Immobilized template recruitment assay: 25 fmol of immobilized G5E4T was preincubated for 20 min at room temperature in the presence (+) and absence (−) of 200 ng GAL4-VP16 with indicated amounts of VP eluate, and/or TFIID (40 ng) and TFIIA (12 ng). After 30 min, unbound protein was removed by washing and the bound protein was resolved on 4%–15% SDS-PAGE. Immunoblots of bound protein are shown. Med130 is used to measure hMed binding; TBP (HA) is representative of TFIID binding. Immunoblots with other subunits of these protein complexes demonstrate identical results.
Figure 6
Preassembly of a complex containing both TFIID and hMed stimulates transcription. (A) Preincubation of TFIID and hMed in the absence of activator enhances basal transcription. The experimental design is outlined at top. The 60-μL reaction mixtures contained 20 ng pG5E4T and, as indicated, 160 ng TFIID, 12 ng TFIIA, or 2 units hMed. Where indicated in the figure, these factors were preincubated (TFIIA was added along with TFIID). Salt and buffer concentrations were held constant throughout the reaction. Thirty-five micrograms of ΔNE, NTPs, and coactivators were added to the preincubation mixes for 4 min at 30°C and transcription was measured by primer extension analysis. An autoradiograph of the gel is shown. (B) Preincubation of TFIID and hMed in the presence of GAL4-VP16 stimulates transcription. All lanes contain 20 ng pG5E4T and 5 ng GAL4-VP16. Lanes 1 and 2 are standard, 30-min in vitro transcription reactions. Addition of 2 units hMed in lane 2 restores activated transcription. Lanes 1 and 2 contain 35 μg mediator-depleted, TFIID-depleted nuclear extract (ΔΔNE) and 2 units hMed. The addition of 160 ng TFIID and 12 ng TFIIA in lane 2 restores activated transcription. ΔΔNE was generated by immunodepletion of TBP, TAFII250, and TAFII32 and is greater than 80-fold depleted of these subunits. The experimental design of preincubation reactions (lanes 3_–_6) is outlined at top. Preincubation, in vitro transcription conditions were the same as those described in panel A with the following exceptions: (1) preincubation was performed in the presence of GAL4-VP16 and 35 μg ΔΔNE; (2) preincubation was for 15 min; (3) the second incubation with remaining factors and NTPs was for 3 min. Shown is a cropped image of an autoradiograph; all lanes are from the same gel.
Figure 6
Preassembly of a complex containing both TFIID and hMed stimulates transcription. (A) Preincubation of TFIID and hMed in the absence of activator enhances basal transcription. The experimental design is outlined at top. The 60-μL reaction mixtures contained 20 ng pG5E4T and, as indicated, 160 ng TFIID, 12 ng TFIIA, or 2 units hMed. Where indicated in the figure, these factors were preincubated (TFIIA was added along with TFIID). Salt and buffer concentrations were held constant throughout the reaction. Thirty-five micrograms of ΔNE, NTPs, and coactivators were added to the preincubation mixes for 4 min at 30°C and transcription was measured by primer extension analysis. An autoradiograph of the gel is shown. (B) Preincubation of TFIID and hMed in the presence of GAL4-VP16 stimulates transcription. All lanes contain 20 ng pG5E4T and 5 ng GAL4-VP16. Lanes 1 and 2 are standard, 30-min in vitro transcription reactions. Addition of 2 units hMed in lane 2 restores activated transcription. Lanes 1 and 2 contain 35 μg mediator-depleted, TFIID-depleted nuclear extract (ΔΔNE) and 2 units hMed. The addition of 160 ng TFIID and 12 ng TFIIA in lane 2 restores activated transcription. ΔΔNE was generated by immunodepletion of TBP, TAFII250, and TAFII32 and is greater than 80-fold depleted of these subunits. The experimental design of preincubation reactions (lanes 3_–_6) is outlined at top. Preincubation, in vitro transcription conditions were the same as those described in panel A with the following exceptions: (1) preincubation was performed in the presence of GAL4-VP16 and 35 μg ΔΔNE; (2) preincubation was for 15 min; (3) the second incubation with remaining factors and NTPs was for 3 min. Shown is a cropped image of an autoradiograph; all lanes are from the same gel.
Figure 7
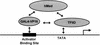
Model of cooperative recruitment of coactivators to DNA templates. Our hypothesis is that reciprocal, cooperative interactions between activator (or series of activators), TFIID, hMed, and template DNA determine the stability of a higher-order nucleoprotein complex. The presence of some form of this higher-order assembly is required for all activated transcription. A key feature of this model is direct communication between the hMed and TFIID.
Similar articles
- The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex.
Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. Drysdale CM, et al. Mol Cell Biol. 1998 Mar;18(3):1711-24. doi: 10.1128/MCB.18.3.1711. Mol Cell Biol. 1998. PMID: 9488488 Free PMC article. - Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s.
Baek HJ, Malik S, Qin J, Roeder RG. Baek HJ, et al. Mol Cell Biol. 2002 Apr;22(8):2842-52. doi: 10.1128/MCB.22.8.2842-2852.2002. Mol Cell Biol. 2002. PMID: 11909976 Free PMC article. - Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly.
Baek HJ, Kang YK, Roeder RG. Baek HJ, et al. J Biol Chem. 2006 Jun 2;281(22):15172-81. doi: 10.1074/jbc.M601983200. Epub 2006 Apr 4. J Biol Chem. 2006. PMID: 16595664 - Mechanisms of viral activators.
Berk AJ, Boyer TG, Kapanidis AN, Ebright RH, Kobayashi NN, Horn PJ, Sullivan SM, Koop R, Surby MA, Triezenberg SJ. Berk AJ, et al. Cold Spring Harb Symp Quant Biol. 1998;63:243-52. doi: 10.1101/sqb.1998.63.243. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384288 Review. - The role of TAFs in RNA polymerase II transcription.
Hahn S. Hahn S. Cell. 1998 Nov 25;95(5):579-82. doi: 10.1016/s0092-8674(00)81625-6. Cell. 1998. PMID: 9845358 Review. No abstract available.
Cited by
- EP400 Deposits H3.3 into Promoters and Enhancers during Gene Activation.
Pradhan SK, Su T, Yen L, Jacquet K, Huang C, Côté J, Kurdistani SK, Carey MF. Pradhan SK, et al. Mol Cell. 2016 Jan 7;61(1):27-38. doi: 10.1016/j.molcel.2015.10.039. Epub 2015 Dec 6. Mol Cell. 2016. PMID: 26669263 Free PMC article. - Genome-wide characterization of Mediator recruitment, function, and regulation.
Grünberg S, Zentner GE. Grünberg S, et al. Transcription. 2017 May 27;8(3):169-174. doi: 10.1080/21541264.2017.1291082. Epub 2017 Feb 8. Transcription. 2017. PMID: 28301289 Free PMC article. Review. - Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation.
Belakavadi M, Fondell JD. Belakavadi M, et al. Mol Cell Biol. 2010 May;30(10):2437-48. doi: 10.1128/MCB.01541-09. Epub 2010 Mar 15. Mol Cell Biol. 2010. PMID: 20231357 Free PMC article. - Molecular and genetic characterization of a Taf1p domain essential for yeast TFIID assembly.
Singh MV, Bland CE, Weil PA. Singh MV, et al. Mol Cell Biol. 2004 Jun;24(11):4929-42. doi: 10.1128/MCB.24.11.4929-4942.2004. Mol Cell Biol. 2004. PMID: 15143185 Free PMC article. - Elongin functions as a loading factor for Mediator at ATF6α-regulated ER stress response genes.
He Y, Sato S, Tomomori-Sato C, Chen S, Goode ZH, Conaway JW, Conaway RC. He Y, et al. Proc Natl Acad Sci U S A. 2021 Sep 28;118(39):e2108751118. doi: 10.1073/pnas.2108751118. Proc Natl Acad Sci U S A. 2021. PMID: 34544872 Free PMC article.
References
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. - PubMed
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. - PubMed
- Carey M, Leatherwood J, Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990a;247:710–712. - PubMed
- Carey M, Lin YS, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990b;345:361–364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases