The structure of a replication initiator unites diverse aspects of nucleic acid metabolism - PubMed (original) (raw)
The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Ramon Campos-Olivas et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Rolling circle replication is a mechanism for copying single-stranded genomes by means of double-stranded intermediates. A multifunctional replication initiator protein (Rep) is indispensable for the precise initiation and termination of this process. Despite the ubiquitous presence and fundamental importance of rolling circle replication elements, structural information on their respective replication initiators is still missing. Here we present the solution NMR structure of the catalytic domain of Rep, the initiator protein of tomato yellow leaf curl virus. It is composed of a central five-stranded anti-parallel beta-sheet, flanked by a small two-stranded beta-sheet, a beta-hairpin and two alpha-helices. Surprisingly, the structure reveals that the catalytic Rep domain is related to a large group of proteins that bind RNA or DNA. Identification of Rep as resembling the family of ribonucleoprotein/RNA-recognition motif fold proteins establishes a structure-based evolutionary link between RNA binding proteins, splicing factors, and replication initiators of prokaryotic and eukaryotic single-stranded DNA elements and mammalian DNA tumor viruses.
Figures
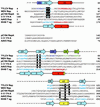
Fig 1.
Domain organization and activity of Rep proteins. (A) Domain structure of TYLCV Rep, wheat dwarf virus (WDV) RepA, porcine circovirus 2 (PCV2) Rep, plasmid pC194 RepA, bacteriophage ΦX174 A, transposon IS91 TnpA, and AAV2 Rep68 proteins. The catalytic domains are displayed in gray, and the conserved sequence motifs (28) are labeled I, II, and III. The oligorimerization domains of TYLCV Rep and WDV RepA are shown in dark gray. (B) Activity assays of TYLCV Rep proteins. (Left) Western blot analysis of a 12% denaturing gel. (Right) A 15% gel after Coomassie staining. About 20–40% of Rep becomes covalently linked to the 5′ end of the 15-nt cleavage product (marked by arrow) under these conditions. The sizes of molecular weight markers are indicated between the panels.
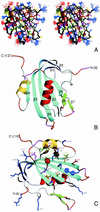
Fig 2.
Three-dimensional structure of TYLCV Rep4–121. (A) Stereoview displaying best-fit superposition of the final ensemble (residues 6 to 119) of 30 conformers with the lowest
dyana
(21) target function (PDB ID ). The protein backbone (N, Cα, CO) is shown in black, and the side chains are colored according to residue type (YFW: brown; D,E: red; K,R,H: blue; A,V,L,I,P: green; T,S,C: yellow; N,Q: magenta). The coordinate precision for the protein backbone heavy atoms is 0.48 Å. (B and C) Ribbon representations of the TYLCV Rep4–121 regularized mean structure (PDB ID ). The central 5-stranded β-sheet is shown in blue, the small extension sheet in dark blue, the helix covering the β-sheet in red, the small 2-stranded sheet in green and loops in gray. The helix carrying the catalytic tyrosine is colored yellow. The strands and helices are numbered and the N and C termini labeled. Loop residues exhibiting substantial flexibility (low 15N heteronuclear NOE) or nondetected NH resonances are colored in orange and magenta, respectively. In C, selected amino acid side chains are displayed as well. They either belong to the conserved sequence motifs or occupy equivalent positions to those implicated in ss- or dsDNA/RNA binding of structurally related proteins (see Fig. 4).
Fig 3.
Structure-based sequence alignment of the catalytic domains of Rep proteins from TYLCV, WDV, pC194, ΦX, AAV2, and the DBD of SV40 T-ag. Amino acids of the motifs I, II, and III (28) are highlighted in black. The catalytic tyrosine(s) and equivalent residues in SV40 T-ag are highlighted in gray. Secondary structure elements present in the TYLCV Rep domain and SV40 T-ag 3D structures are indicated by cylinders (α-helices) and arrows (β-strands) above and below the sequence alignment, respectively. Amino acids in β-strands and α-helices of TYLCV Rep and SV40 T-ag, as well as predicted ones, are colored according to their location in the structure (see Fig. 2_B_ and C). Residue numbers are given at the end of each line. (GenBank accession nos: TYLCV Rep, ; WDV RepA, ; PCV2 Rep, ; pC194 RepA, NP_040435; IS91 TnpA, ; ΦX174 A, NP_040703; AAV2 Rep68, ; SV40 T-ag, ).
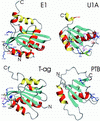
Fig 4.
Ribbon representation of the DBDs of papillomavirus protein E1 and SV40 T-ag and RBDs of U1A and PTB. Helices conserved between Rep and D/RBDs are colored in red (α1) and yellow (α2), all other helices in red/yellow (outside/inside), strands in blue and loops and chain termini in gray. Side chains of amino acids implicated in nucleic acid recognition are displayed and colored identically to the color code given for Fig. 2_C_. PDB accession codes are E1, ; U1A, ; T-ag, ; and PTB, .
Similar articles
- DNA-binding specificity determinants of replication proteins encoded by eukaryotic ssDNA viruses are adjacent to widely separated RCR conserved motifs.
Londoño A, Riego-Ruiz L, Argüello-Astorga GR. Londoño A, et al. Arch Virol. 2010 Jul;155(7):1033-46. doi: 10.1007/s00705-010-0674-4. Epub 2010 Apr 27. Arch Virol. 2010. PMID: 20422235 - Functional mapping of the DNA binding domain of bovine papillomavirus E1 protein.
West M, Flanery D, Woytek K, Rangasamy D, Wilson VG. West M, et al. J Virol. 2001 Dec;75(24):11948-60. doi: 10.1128/JVI.75.24.11948-11960.2001. J Virol. 2001. PMID: 11711585 Free PMC article. - Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids.
Giraldo R, Fernández-Tresguerres ME. Giraldo R, et al. Plasmid. 2004 Sep;52(2):69-83. doi: 10.1016/j.plasmid.2004.06.002. Plasmid. 2004. PMID: 15336485 Review. - Mitochondrial single-stranded DNA-binding proteins: in search for new functions.
Tomáska L, Nosek J, Kucejová B. Tomáska L, et al. Biol Chem. 2001 Feb;382(2):179-86. doi: 10.1515/BC.2001.025. Biol Chem. 2001. PMID: 11308016 Review.
Cited by
- Metal-Induced Stabilization and Activation of Plasmid Replication Initiator RepB.
Ruiz-Masó JA, Bordanaba-Ruiseco L, Sanz M, Menéndez M, Del Solar G. Ruiz-Masó JA, et al. Front Mol Biosci. 2016 Sep 21;3:56. doi: 10.3389/fmolb.2016.00056. eCollection 2016. Front Mol Biosci. 2016. PMID: 27709114 Free PMC article. - The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.
Kosek D, Grabundzija I, Lei H, Bilic I, Wang H, Jin Y, Peaslee GF, Hickman AB, Dyda F. Kosek D, et al. Mol Cell. 2021 Oct 21;81(20):4271-4286.e4. doi: 10.1016/j.molcel.2021.07.028. Epub 2021 Aug 16. Mol Cell. 2021. PMID: 34403695 Free PMC article. - A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue.
de Villiers EM, Gunst K, Chakraborty D, Ernst C, Bund T, Zur Hausen H. de Villiers EM, et al. Emerg Microbes Infect. 2019;8(1):1205-1218. doi: 10.1080/22221751.2019.1651620. Emerg Microbes Infect. 2019. PMID: 31409221 Free PMC article. - CRISPR-Cas provides limited phage immunity to a prevalent gut bacterium in gnotobiotic mice.
Rasmussen TS, Koefoed AK, Deng L, Muhammed MK, Rousseau GM, Kot W, Sprotte S, Neve H, Franz CMAP, Hansen AK, Vogensen FK, Moineau S, Nielsen DS. Rasmussen TS, et al. ISME J. 2023 Mar;17(3):432-442. doi: 10.1038/s41396-023-01358-4. Epub 2023 Jan 11. ISME J. 2023. PMID: 36631688 Free PMC article. - Nuclear targeting of a bacterial integrase that mediates site-specific recombination between bacterial and human target sequences.
Agúndez L, Machón C, César CE, Rosa-Garrido M, Delgado MD, Llosa M. Agúndez L, et al. Appl Environ Microbiol. 2011 Jan;77(1):201-10. doi: 10.1128/AEM.01371-10. Epub 2010 Oct 29. Appl Environ Microbiol. 2011. PMID: 21037296 Free PMC article.
References
- Kornberg A. & Baker, T. A., (1992) DNA Replication (Freeman, New York).
- Been M. D. & Wickham, G. S. (1997) Eur. J. Biochem. 247 741-753. - PubMed
- Khan S. A. (2000) Mol. Microbiol. 37 477-484. - PubMed
- Baas P. D. & Jansz, H. S. (1988) Curr. Top. Microbiol. Immunol. 136 31-70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources