A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5' end of the latency-associated transcript produces a protein in infected rabbits - PubMed (original) (raw)
. 2002 Aug;76(16):8003-10.
doi: 10.1128/jvi.76.16.8003-8010.2002.
Barak Maguen, Ling Jin, Kevin R Mott, John Kurylo, Lbachir BenMohamed, Ada Yukht, Nelson Osorio, Anthony B Nesburn, Gail Henderson, Melissa Inman, Clinton Jones, Steven L Wechsler
Affiliations
- PMID: 12134005
- PMCID: PMC155148
- DOI: 10.1128/jvi.76.16.8003-8010.2002
A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5' end of the latency-associated transcript produces a protein in infected rabbits
Guey-Chuen Perng et al. J Virol. 2002 Aug.
Abstract
Following primary ocular infection, herpes simplex virus type 1 (HSV-1) establishes a lifelong latent infection in sensory neurons of the trigeminal ganglia. Latency-associated transcript (LAT), the only known viral gene abundantly transcribed during HSV-1 neuronal latency, is required for high levels of reactivation. Recently we showed that three different mutants that do not alter the LAT promoter but contain deletions within the 5' end of the primary LAT transcript affect viral virulence (G. C. Perng et al., J. Virol. 75:9018-9028, 2001). In contrast, in LAT-null mutants viral virulence appears unaltered (T. M. Block et al., Virology 192:618-630, 1993; D. C. Bloom et al., J. Virol. 68:1283-1292, 1994; J. M. Hill et al., Virology 174:117-125, 1990; G. C. Perng et al., J. Virol. 68:8045-8055, 1994; F. Sedarati, K. M. Izumi, E. K. Wagner, and J. G. Stevens, J. Virol. 63:4455-4458, 1989). We therefore hypothesized that the 5' end of LAT and/or an as yet unidentified gene that overlaps part of this region is involved in viral virulence. We report here on the discovery and initial characterization of a novel HSV-1 RNA consistent with such a putative gene. The novel RNA was antisense to the 5' end of LAT and was designated AL-RNA (anti-LAT sense RNA). The AL-RNA overlapped the core LAT promoter and the first 158 nucleotides of the 5' end of the primary LAT transcript. AL-RNA was detected in extracts from neuron-like cells (PC-12) infected with wild-type HSV-1 but not in cells infected with a mutant with the AL region deleted. The deletions in each of the above three mutants with altered virulence encompass the 5' end of the AL-RNA, and these mutants cannot transcribe AL. This supports the hypothesis that the AL gene may play a role in viral virulence. Based on comparison to the corresponding genomic sequence, the AL-RNA did not appear to be spliced. The AL-RNA was polyadenylated and contained an open reading frame capable of encoding a protein 56 amino acids in length with a predicted molecular mass of 6.8 kDa. Sera from three of three rabbits infected with wild-type HSV-1 but not sera from any of three rabbits infected with a mutant with the AL-RNA region deleted recognized the Escherichia coli recombinantly expressed AL open reading frame on Western blots. In addition, four of six rabbits infected with wild-type virus developed enzyme-linked immunosorbent assay titers against one or more AL synthetic peptides. These results suggest that an AL protein is produced in vivo.
Figures
FIG. 1.
Relative location of the AL gene. (A) Schematic representation of the wild-type HSV-1 genome. TRL and IRL indicate the terminal and inverted long repeats, respectively. IRS and TRS indicate the inverted and terminal short repeats, respectively. UL and US indicate the unique long and unique short regions, respectively. The TRL and IRL are expanded, and the TRL is flipped left to right as indicated by the dashed lines, so that both repeats can be represented together in the subsequent panels. (B) Blowup of the long repeats. The primary LAT transcript is indicated by the large arrow. The solid rectangle represents the very stable 2-kb LAT intron. The LAT TATA box is indicated by TATA. The start of LAT transcription is indicated by the arrow at +1 (genomic nucleotide 118801). Several restriction enzyme sites and the relative locations of the ICP0 and ICP34.5 transcripts are shown for reference. (C) Blowup of the 5′ LAT region. The LAT promoter (open rectangle), the start of LAT transcription (arrow, +1), a putative secondary LAT promoter (LAP2), and the beginning of the stable 2-kb LAT are indicated. Nucleotide positions relative to the start of LAT transcription are shown in parentheses. (D) AL gene. The DNA strand opposite that of LAT in panel C is shown. The numbers in parentheses indicate nucleotide positions relative to the start of LAT transcription on the other strand. The remaining numbers indicate nucleotide positions relative to the start of AL transcription. The ORF encoding a putative AL protein is shown as an open rectangle. The AL promoter is located somewhere between the large parentheses in the same region as LAP2. (E) Relative positions of oligonucleotide primers (a to d) and probes (e) used in this report. a, 118661 to 118690; b, 118681 to 118710; c, 118841 to 118870; d, 118881 to 118910; e, 118811 to 118840
FIG. 2.
Detection of AL-RNA by RT-PCR. PC-12 cells were infected at an MOI of 5. Total RNA was isolated 6 h p.i., and RT-PCR was performed as described in Materials and Methods. Lanes: 1, no RT, wild-type McKrae virus-infected PC-12 cells; 2, wild-type McKrae virus; 3, dLAT2903R; 4, dLAT2903; 5, uninfected PC-12; 6, no RT, dLAT2903; 7, no RT, uninfected PC-12; 8, marker, PCR of wild-type McKrae viral DNA.
FIG. 3.
Mapping the 3′ end of the AL-RNA. RT-PCR was performed on poly(A)+ RNA isolated from PC-12 cells infected with wild-type McKrae virus, and the RT-PCR product was cloned and sequenced as described in Materials and Methods. The resulting sequence is labeled RNA. The “genome” sequence is from the same HSV-1 strain (McKrae). The asterisks in the “genome” sequence indicate identity to the RNA sequence. This identity ends after genomic nucleotide 118603. The positions of the 5′ primer and the 3′ oligo(dT) primer are shown.
FIG. 4.
Kinetics of AL-RNA expression. PC-12 cells were infected at an MOI of 5 with wild-type McKrae virus. Total RNA was isolated at various times p.i., and RT-PCR was performed as described in Materials and Methods. Lanes: 1, positive control (PCR of plasmid AL DNA); 2, uninfected; 3, 2 h p.i.; 4, 4 h p.i.; 5, 6 h p.i.; 6, 8 h p.i.; 7, 10 h p.i.; 8, 12 h p.i.; 9, 14 h p.i.; 10, no RT, 8 h p.i.
FIG. 5.
Complete sequence of AL-RNA. The sequence was determined as described in the text. AL-RNA indicates the DNA sequence corresponding to the AL-RNA sequence. The genomic sequence is from McKrae (the same HSV-1 strain as the RNA). The AL ORF begins with a methionine (M) at AL nucleotide 61 and ends with a TAG at AL nucleotide 229. The boxed C at genomic nucleotide location 118769 indicates the only nucleotide difference in AL between McKrae and HSV-1 strain 17syn+, which contains a G at this location. The boxed Q indicates that the nucleotide change from C to G changed the predicted amino acid from Q in McKrae to E in 17syn+.
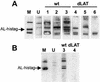
FIG. 6.
Detection of _E. coli_-expressed AL protein by sera from infected rabbits. (A) Total extract from E. coli expressing the AL-His tag fusion protein was run on a 15% Tricine gel with a single large loading well and transferred to a PVDF membrane. The membrane was cut into strips, and each strip was separately reacted with the indicated antibody. The strips were then reacted with horseradish peroxidase-conjugated secondary antibody for chemifluorescence. The arrow indicates the location of the _E. coli_-expressed AL-His tag fusion protein. Lane M, anti-His tag antibody as marker. Lane U, serum from an uninfected rabbit. Lanes 1 to 3, sera from three different rabbits infected with wild-type virus. Lanes 4 to 6, sera from three different rabbits infected with dLAT2903 (a LAT and AL-null mutant). (B) The _E. coli_-expressed AL-His tag protein was partially purified as described in Materials and Methods and processed as for panel A. Lane M, anti-His tag antibody as marker. Lane U, serum from an uninfected rabbit different from the serum in panel A. Lanes 3 and 4, the same rabbit sera as in lanes 3 and 4 in panel A. wt, wild type.
Similar articles
- A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels.
Perng GC, Maguen B, Jin L, Mott KR, Osorio N, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, Inman M, Henderson G, Jones C, Wechsler SL. Perng GC, et al. J Virol. 2002 Feb;76(3):1224-35. doi: 10.1128/jvi.76.3.1224-1235.2002. J Virol. 2002. PMID: 11773398 Free PMC article. - Identification of a protein encoded in the herpes simplex virus type 1 latency associated transcript promoter region.
Naito J, Mukerjee R, Mott KR, Kang W, Osorio N, Fraser NW, Perng GC. Naito J, et al. Virus Res. 2005 Mar;108(1-2):101-10. doi: 10.1016/j.virusres.2004.08.011. Virus Res. 2005. PMID: 15681060 - The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript.
Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. Perng GC, et al. J Virol. 1996 Feb;70(2):976-84. doi: 10.1128/JVI.70.2.976-984.1996. J Virol. 1996. PMID: 8551638 Free PMC article. - HSV LAT and neuronal survival.
Bloom DC. Bloom DC. Int Rev Immunol. 2004 Jan-Apr;23(1-2):187-98. doi: 10.1080/08830180490265592. Int Rev Immunol. 2004. PMID: 14690860 Review.
Cited by
- Blockade of LAG-3 Immune Checkpoint Combined With Therapeutic Vaccination Restore the Function of Tissue-Resident Anti-viral CD8+ T Cells and Protect Against Recurrent Ocular Herpes Simplex Infection and Disease.
Roy S, Coulon PG, Srivastava R, Vahed H, Kim GJ, Walia SS, Yamada T, Fouladi MA, Ly VT, BenMohamed L. Roy S, et al. Front Immunol. 2018 Dec 17;9:2922. doi: 10.3389/fimmu.2018.02922. eCollection 2018. Front Immunol. 2018. PMID: 30619285 Free PMC article. - Hidden regulation of herpes simplex virus 1 pre-mRNA splicing and polyadenylation by virally encoded immediate early gene ICP27.
Tang S, Patel A, Krause PR. Tang S, et al. PLoS Pathog. 2019 Jun 17;15(6):e1007884. doi: 10.1371/journal.ppat.1007884. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31206552 Free PMC article. - Identification of a novel bovine herpesvirus 1 transcript containing a small open reading frame that is expressed in trigeminal ganglia of latently infected cattle.
Inman M, Zhou J, Webb H, Jones C. Inman M, et al. J Virol. 2004 May;78(10):5438-47. doi: 10.1128/jvi.78.10.5438-5447.2004. J Virol. 2004. PMID: 15113922 Free PMC article. - A novel HLA (HLA-A*0201) transgenic rabbit model for preclinical evaluation of human CD8+ T cell epitope-based vaccines against ocular herpes.
Chentoufi AA, Dasgupta G, Christensen ND, Hu J, Choudhury ZS, Azeem A, Jester JV, Nesburn AB, Wechsler SL, BenMohamed L. Chentoufi AA, et al. J Immunol. 2010 Mar 1;184(5):2561-71. doi: 10.4049/jimmunol.0902322. Epub 2010 Feb 1. J Immunol. 2010. PMID: 20124097 Free PMC article. - The latency-associated transcript locus of herpes simplex virus 1 is a virulence determinant in human skin.
Vanni EAH, Foley JW, Davison AJ, Sommer M, Liu D, Sung P, Moffat J, Zerboni L, Arvin AM. Vanni EAH, et al. PLoS Pathog. 2020 Dec 28;16(12):e1009166. doi: 10.1371/journal.ppat.1009166. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33370402 Free PMC article.
References
- Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618-630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY012823/EY/NEI NIH HHS/United States
- R01 EY013191/EY/NEI NIH HHS/United States
- EY11629/EY/NEI NIH HHS/United States
- EY13191/EY/NEI NIH HHS/United States
- EY12823/EY/NEI NIH HHS/United States
- P20RR15635/RR/NCRR NIH HHS/United States
- EY07566/EY/NEI NIH HHS/United States
- P20 RR015635/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials