Molecular anatomy of the human excision nuclease assembled at sites of DNA damage - PubMed (original) (raw)
Molecular anatomy of the human excision nuclease assembled at sites of DNA damage
Joyce T Reardon et al. Mol Cell Biol. 2002 Aug.
Abstract
Human nucleotide excision repair is initiated by six repair factors (XPA, RPA, XPC-HR23B, TFIIH, XPF-ERCC1, and XPG) which sequentially assemble at sites of DNA damage and effect excision of damage-containing oligonucleotides. We here describe the molecular anatomy of the human excision nuclease assembled at the site of a psoralen-adducted thymine. Three polypeptides, primarily positioned 5' to the damage, are in close physical proximity to the psoralen lesion and thus are cross-linked to the damaged DNA: these proteins are RPA70, RPA32, and the XPD subunit of TFIIH. While both XPA and XPC bind damaged DNA and are required for XPD cross-linking to the psoralen-adducted base, neither XPA nor XPC is cross-linked to the psoralen adduct. The presence of other repair factors, in particular TFIIH, alters the mode of RPA binding and the position of its subunits relative to the psoralen lesion. Based on these results, we propose that RPA70 makes the initial contact with psoralen-damaged DNA but that within preincision complexes, it is RPA32 and XPD that are in close contact with the lesion.
Figures
FIG. 1.
DNA substrates used in this study. The sequence of the 140-bp duplex with a psoralen-adducted thymine at position 70 (underlined) is shown in panel A. The arrows above and below the sequence indicate the ligation sites between the six oligomers used to assemble the substrate. Radiolabels were introduced in the top strand at the sixth phosphodiester bond 5′ to the lesion or at the seventh phosphodiester bond 3′ to the adduct to generate 5′ and 3′ labeled substrates, respectively. The duplex contained a furan-side monoadduct, and the chemical structure of HMT is shown in panel B. The HMT adduct locally disrupts the helical structure (39), and TFIIH further unwinds the duplex in both the 5′ and 3′ directions as illustrated in panel C (adapted from reference 25).
FIG. 2.
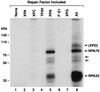
RPA70 and RPA32 are photo-cross-linked to psoralen-damaged DNA. The substrate was a 140-bp duplex with 32P radiolabel six phosphodiester bonds 5′ to the psoralen-adducted thymine (Fig. 1). The indicated repair factors (at the same concentrations used to reconstitute excision repair) were incubated with DNA for 60 min and irradiated with black light, and then reaction mixtures were digested with DNase I. Following resolution in a SDS-10% polyacrylamide gel, DNA-protein complexes were visualized by autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands (indicated with open arrows) migrating between RPA70-DNA and RPA32-DNA complexes result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitative analyses. The most slowly migrating complex, designated (XPD), was identified as an XPD-DNA complex in subsequent experiments.
FIG. 3.
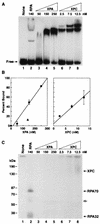
XPA and XPC bind DNA but are not cross-linked to psoralen. Panel A shows an autoradiograph of DNA-protein complexes resolved in a nondenaturing polyacrylamide gel. Excision repair is typically reconstituted with 140 nM RPA, 50 nM XPA, and 2.5 nM XPC (lanes 2, 3, and 6). For quantitation, the fraction of DNA not in the gel position corresponding to free DNA was considered bound. The average percent binding for this experiment and a second electrophoretic mobility shift assay conducted under the same conditions are shown in panel B; in the plot on the left, RPA-DNA binding is indicated by the filled triangle. Panel C illustrates the results of a photo-cross-linking experiment performed with half of the same reactions shown in panel A; to the left are positions of molecular mass markers resolved in the same gel. The fainter band (indicated with an open arrow) migrating between RPA70-DNA and RPA32-DNA complexes results from cross-linking proteolytic fragments of RPA70 to the substrate and was not included in quantitative analyses.
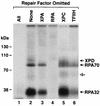
FIG. 4.
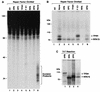
Single-factor omission experiments for excision repair and cross-linking assays reveal that the third complex contains XPD. Repair factors (at the same concentrations used to reconstitute excision repair), with the omitted factor indicated, were incubated with psoralen-damaged DNA for 60 min and then either processed for detection of excised oligomers (A) or photo-cross-linked to detect DNA-protein complexes (B). To detect excision, deproteinized DNA was resolved in a 10% sequencing gel with _Hin_fI digested DNA as size markers (mobility positions, in nucleotides, are shown to the left) and visualized by autoradiography (A). To detect DNA-protein complexes, reaction mixtures were resolved in an SDS-8% polyacrylamide gel with size markers (indicated to the left) and visualized by autoradiography (B). Only the portion of the gel corresponding to protein-DNA complexes greater than approximately 70 kDa is shown. Panel C illustrates in vitro-labeled proteins resolved in an SDS-10% polyacrylamide gel with a complete repair reaction that was cross-linked and treated with DNase I; only the relevant section of the gel is shown to illustrate the migration of the TFIIH and RPA70 complexes (lane 4) relative to 35S-labeled XPB, RPA70, and XPD proteins (lanes 1 to 3).
FIG. 5.
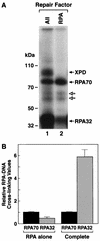
RPA subunits are differentially cross-linked when incubated alone or in combination with other repair factors. RPA was incubated with substrate DNA as described previously either alone or with other repair factors (at the same concentrations used to reconstitute excision repair) for 60 min and irradiated with black light, and then reaction mixtures were digested with DNase I. Following resolution in an SDS-10% polyacrylamide gel, the DNA-protein complexes were visualized by autoradiography (A). Lane 1 is a complete reaction, and lane 2 is RPA alone; to the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands migrating between RPA70-DNA and RPA32-DNA complexes (indicated with open arrows) result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitation. Panel B is a graphic summarization of relative RPA70/RPA32 cross-linking values for RPA alone reactions (n = 8) and complete reactions (n = 16). Average values are plotted, and error bars represent the standard error. For each data set the relative RPA70 signal is defined as 1.
FIG. 6.
Single-factor omission experiments for photo-cross-linking reveal that inversion of the RPA70/RPA32 ratio is dependent on TFIIH. The substrate was a 140-bp duplex with 32P radiolabel six phosphodiester bonds 5′ to the psoralen-adducted thymine (Fig. 1). The indicated repair factors (at the same concentrations used to reconstitute excision repair) were incubated with DNA for 60 min and irradiated with black light, and reaction mixtures were digested with DNase I. Following resolution in an SDS-10% polyacrylamide gel, DNA-protein complexes were visualized by autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter band (indicated with an open arrow) migrating between RPA70-DNA and RPA32-DNA complexes results from cross-linking proteolytic fragments of RPA70 to the substrate and was not included in quantitative analyses.
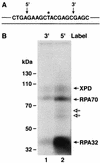
FIG. 7.
Repair factor binding is preferentially located 5′ to the psoralen damage. Panel A is a schematic diagram of the psoralen 140-bp duplex, highlighting the central 20 nt; the substrate extends 60 nt in both the 5′ and 3′ directions, and the locations of 32P radiolabel are indicated by arrows above the sequence. Panel B is an autoradiograph of an SDS-10% polyacrylamide gel. The complete complement of repair factors was incubated with either the 3′ labeled substrate (lane 1) or the 5′ labeled substrate (lane 2), irradiated with black light, and reaction mixtures were digested with DNase I prior to electrophoresis and autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands migrating between RPA70-DNA and RPA32-DNA complexes (indicated by open arrows) result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitative analyses.
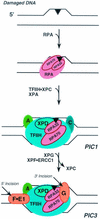
FIG. 8.
Molecular anatomy of preincision complexes. DNA damage, represented by a filled triangle, often disrupts base pairing in the immediate vicinity of the adduct. RPA subunits, designated RPA70 and RPA32, contact DNA damage in the absence of other repair factors, and the RPA70 subunit is preferentially photo-cross-linked to the psoralen lesion. The first stable preincision complex (PIC1) contains XPA (A), RPA (for clarity only the two larger subunits are shown), XPC-HR23B (C), and the multimeric TFIIH (for clarity only the XPD subunit is shown). In PIC1 TFIIH further unwinds the helix ∼20 nt in the region of DNA damage, RPA undergoes a conformational change, and the relative cross-linking of RPA subunits is inverted such that RPA32 is preferentially photo-cross-linked; this preferential cross-linking is also observed in PIC2 and PIC3. In PIC1 the XPD subunit of TFIIH is also in contact with the lesion, and XPD remains in contact with psoralen in PIC2 and PIC3. The ultimate preincision complex (PIC3) is also shown: XPC-HR23B is replaced by XPG (G) 3′ to the lesion, XPF-ERCC1 (F•E1) assembles 5′ to the damage, and dual incisions and excision (indicated by arrows) occur.
Similar articles
- Mechanism of open complex and dual incision formation by human nucleotide excision repair factors.
Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Evans E, et al. EMBO J. 1997 Nov 3;16(21):6559-73. doi: 10.1093/emboj/16.21.6559. EMBO J. 1997. PMID: 9351836 Free PMC article. - Order of assembly of human DNA repair excision nuclease.
Wakasugi M, Sancar A. Wakasugi M, et al. J Biol Chem. 1999 Jun 25;274(26):18759-68. doi: 10.1074/jbc.274.26.18759. J Biol Chem. 1999. PMID: 10373492 - Novel functional interactions between nucleotide excision DNA repair proteins influencing the enzymatic activities of TFIIH, XPG, and ERCC1-XPF.
Winkler GS, Sugasawa K, Eker AP, de Laat WL, Hoeijmakers JH. Winkler GS, et al. Biochemistry. 2001 Jan 9;40(1):160-5. doi: 10.1021/bi002021b. Biochemistry. 2001. PMID: 11141066 - DNA damage recognition during nucleotide excision repair in mammalian cells.
Wood RD. Wood RD. Biochimie. 1999 Jan-Feb;81(1-2):39-44. doi: 10.1016/s0300-9084(99)80036-4. Biochimie. 1999. PMID: 10214908 Review. - XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.
Fuss JO, Tainer JA. Fuss JO, et al. DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review.
Cited by
- Expression and Clinical Significance of ERCC1 and XPF in Human Hepatocellular Carcinoma.
Liao X, Li Y, Li H, Huang W, Wang H, Xie W. Liao X, et al. Onco Targets Ther. 2020 Feb 4;13:1059-1072. doi: 10.2147/OTT.S237916. eCollection 2020. Onco Targets Ther. 2020. PMID: 32099408 Free PMC article. - Polymorphisms of DNA repair genes are associated with colorectal cancer in patients with Lynch syndrome.
Kamiza AB, Hsieh LL, Tang R, Chien HT, Lai CH, Chiu LL, Lo TP, Hung KY, You JF, Wang WC, Hsiung CA, Yeh CC. Kamiza AB, et al. Mol Genet Genomic Med. 2018 Apr 17;6(4):533-40. doi: 10.1002/mgg3.402. Online ahead of print. Mol Genet Genomic Med. 2018. PMID: 29664240 Free PMC article. - Analysis of DNA binding by human factor xeroderma pigmentosum complementation group A (XPA) provides insight into its interactions with nucleotide excision repair substrates.
Sugitani N, Voehler MW, Roh MS, Topolska-Woś AM, Chazin WJ. Sugitani N, et al. J Biol Chem. 2017 Oct 13;292(41):16847-16857. doi: 10.1074/jbc.M117.800078. Epub 2017 Aug 31. J Biol Chem. 2017. PMID: 28860187 Free PMC article. - Photoaffinity labeling of transcription factors by DNA-templated crosslinking.
Liu Y, Zheng W, Zhang W, Chen N, Liu Y, Chen L, Zhou X, Chen X, Zheng H, Li X. Liu Y, et al. Chem Sci. 2015 Jan 1;6(1):745-751. doi: 10.1039/c4sc01953a. Epub 2014 Oct 1. Chem Sci. 2015. PMID: 28706637 Free PMC article. - PostExcision Events in Human Nucleotide Excision Repair.
Kemp MG, Hu J. Kemp MG, et al. Photochem Photobiol. 2017 Jan;93(1):178-191. doi: 10.1111/php.12641. Epub 2016 Oct 27. Photochem Photobiol. 2017. PMID: 27645806 Free PMC article. Review.
References
- Chang, W.-H., and R. D. Kornberg. 2000. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell 102:609-613. - PubMed
- Cimino, G. D., H. B. Gamper, S. T. Isaacs, and J. E. Hearst. 1985. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 54:1151-1193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials