DksA affects ppGpp induction of RpoS at a translational level - PubMed (original) (raw)
DksA affects ppGpp induction of RpoS at a translational level
Larissa Brown et al. J Bacteriol. 2002 Aug.
Abstract
The RpoS sigma factor (also called sigmaS or sigma38) is known to regulate at least 50 genes in response to environmental sources of stress or during entry into stationary phase. Regulation of RpoS abundance and activity is complex, with many factors participating at multiple levels. One factor is the nutritional stress signal ppGpp. The absence of ppGpp blocks or delays the induction of rpoS during entry into stationary phase. Artificially inducing ppGpp, without starvation, is known to induce rpoS during the log phase 25- to 50-fold. Induction of ppGpp is found to have only minor effects on rpoS transcript abundance or on RpoS protein stability; instead, the efficiency of rpoS mRNA translation is increased by ppGpp as judged by both RpoS pulse-labeling and promoter-independent effects on lacZ fusions. DksA is found to affect RpoS abundance in a manner related to ppGpp. Deleting dksA blocks rpoS induction by ppGpp. Overproduction of DksA induces rpoS but not ppGpp. Deleting dksA neither alters regulation of ppGpp in response to amino acid starvation nor nullifies the inhibitory effects of ppGpp on stable RNA synthesis. Although this suggests that dksA is epistatic to ppGpp, inducing ppGpp does not induce DksA. A dksA deletion does display a subset of the same multiple-amino-acid requirements found for ppGpp(0) mutants, but overproducing DksA does not satisfy ppGpp(0) requirements. Sequenced spontaneous extragenic suppressors of dksA polyauxotrophy are frequently the same T563P rpoB allele that suppresses a ppGpp(0) phenotype. We propose that DksA functions downstream of ppGpp but indirectly regulates rpoS induction.
Figures
FIG. 1.
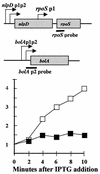
Overproduction of ppGpp does not stimulate rpoS transcription. Upper panel, the rpoS and bolA operons are shown schematically with black bars indicating mRNA regions hybridizing to RNA probes. Graph, activities measured at various times in minutes are normalized to an actin RNA external standard. Filled symbols = rpoS probe hybrids. Open symbols = bolA probe hybrids. Induction of ppGpp was achieved by adding IPTG to exponentially growing cells bearing plasmid pALS13 to overexpress a fragment of the RelA protein that is constitutive for ppGpp synthetic activity.
FIG. 2.
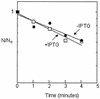
Overproduction of ppGpp does not affect RpoS stability. Metabolic decay of pulse-labeled RpoS was measured during a chase imposed either before (open squares) or 10 min after (closed circles) induction of ppGpp. The apparent rates of exponential decay of activities lead to estimates of the RpoS half-life as about 5 min before and 6 min after IPTG induction.
FIG. 3.
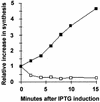
Overproduction of ppGpp increases specific rates of RpoS synthesis while decreasing overall rates of protein synthesis. At times indicated after IPTG addition, samples were pulse-labeled with [35S]Met/Cys for 30 and 60 s and initial rates of synthesis (plotted) were determined from the slopes. Activities associated with RpoS by SDS-PAGE of immunoprecipitates were used to estimate a rate of RpoS synthesis for each induction time normalized to rates of synthesis before IPTG addition (closed squares). Total protein synthesis rates (open squares) were estimated from the slope obtained for total TCA-precipitable activity recovered on filters after both pulses without immunoprecipitation, normalized to rates before IPTG addition.
FIG. 4.
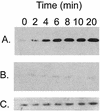
A dksA mutation impairs induction of RpoS by overproduction of ppGpp in exponential phase. Cultures were grown in LB medium to an A_600 of 0.3 when IPTG was added to induce ppGpp (time zero), and timed samples were assayed for RpoS by Western blotting. (A) CF1648 (wild type/pALS13). (B) TE8114 (Δ_dksA/pALS13). (C) Overexposure of the blot in panel B in order to visualize the small amount of RpoS detected.
FIG. 5.
RpoS induction during growth into stationary phase is affected by DksA. Left, LB culture growth. Samples 1 to 9 were taken at the points indicated by the arrows, while sample 10 represents an overnight culture. Right, RpoS Western blots of cultures from top to bottom: wild type (MG1655); dksA_− (TE8114 = MG1655 but dksA::tet); dksA_−/p_dksA (TE8114/pJK537); ppGpp0 (CF1693 = MG1655 but Δ_relA Δ_spoT_); and ppGpp0/p_dksA_ (CF1693/pJK537).
FIG. 6.
DksA does not alter ppGpp regulation during the stringent response. Cultures were grown and uniformly labeled with 32Pi in MOPS minimal glucose medium, five nucleosides, and all amino acids except serine. At time zero, 1 mg of serine hydroxamate/ml was added and the culture was assayed for guanine nucleotide content at various times thereafter. The amount of ppGpp is normalized to the sum of GTP, ppGpp, and pppGpp.
FIG. 7.
The presence or absence of ppGpp does not alter the DksA accumulation. Cultures of strains A to D were grown in LB medium, and in each case six samples were taken representing the growth transition to stationary phase as in Fig. 5. The DksA content was analyzed by Western blots. (A) CF1648 = MG1655 (wild type); (B) CF1648/p_dksA_; (C) CF1693 Δ_relA_ Δ_spoT_ ppGpp0; and (D) CF1693/p_dksA_. The presence of the p_dksA_ plasmid is indicated by a plus at the top of sample series B and D.
FIG. 8.
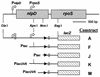
Schematics of rpoS-lac fusions. The top line shows diagrams of the nlpD and rpoS genes of E. coli. Native rpoS is transcribed from two promoters indicated by the bent arrows. The lower lines represent the different protein fusions made by Cunning et al. (11) that were transduced into an rph+ MG1655 derivative with an internal lacIZ deletion (CF7968) for analysis. The AUG start codons are indicated by open squares, and stop codons are indicated by arrowheads.
FIG. 9.
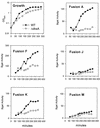
β-Galactosidase assays of rpoS-lac fusions in a dksA deletion mutant during growth into stationary phase. (A) Cultures were grown in MOPS minimal media with glucose, all amino acids, and the five nucleosides. Upper left panel (Growth) shows typical growth for each of the different fusions. Remaining labeled panels show β-galactosidase-specific activities for each fusion construct assayed. OD600, optical density at 600 nm.
FIG. 10.
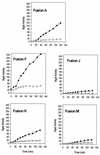
β-Galactosidase assays of rpoS-lac fusions after induction of ppGpp. Cultures were grown in LB medium, and 0.2% arabinose was added at an A_600 of 0.3 (time zero) to induce relA expression from pRelA. Samples thereafter were assayed for β-galactosidase. Specific activities for each fusion are shown. Filled circles, wild type; and open circles, Δ_dksA.
Similar articles
- Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli.
Hirsch M, Elliott T. Hirsch M, et al. J Bacteriol. 2002 Sep;184(18):5077-87. doi: 10.1128/JB.184.18.5077-5087.2002. J Bacteriol. 2002. PMID: 12193624 Free PMC article. - DksA and ppGpp Regulate the σS Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP.
Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. Girard ME, et al. J Bacteriol. 2017 Dec 20;200(2):e00463-17. doi: 10.1128/JB.00463-17. Print 2018 Jan 15. J Bacteriol. 2017. PMID: 29061665 Free PMC article. - Control of bacterial transcription, translation and replication by (p)ppGpp.
Srivatsan A, Wang JD. Srivatsan A, et al. Curr Opin Microbiol. 2008 Apr;11(2):100-5. doi: 10.1016/j.mib.2008.02.001. Epub 2008 Mar 24. Curr Opin Microbiol. 2008. PMID: 18359660 Review. - Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity.
Sharma UK, Chatterji D. Sharma UK, et al. FEMS Microbiol Rev. 2010 Sep;34(5):646-57. doi: 10.1111/j.1574-6976.2010.00223.x. Epub 2010 Apr 14. FEMS Microbiol Rev. 2010. PMID: 20491934 Review.
Cited by
- Bacterial cross-feeding can promote gene retention by lowering gene expression costs.
Chuang YC, Behringer MG, Patton G, Bird JT, Love CE, Dalia A, McKinlay JB. Chuang YC, et al. bioRxiv [Preprint]. 2024 Aug 19:2024.08.19.608702. doi: 10.1101/2024.08.19.608702. bioRxiv. 2024. PMID: 39229193 Free PMC article. Preprint. - Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli.
Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. Magnusson LU, et al. J Bacteriol. 2007 Jul;189(14):5193-202. doi: 10.1128/JB.00330-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496080 Free PMC article. - The RpoS-mediated general stress response in Escherichia coli.
Battesti A, Majdalani N, Gottesman S. Battesti A, et al. Annu Rev Microbiol. 2011;65:189-213. doi: 10.1146/annurev-micro-090110-102946. Annu Rev Microbiol. 2011. PMID: 21639793 Free PMC article. Review. - Role of a Zn-independent DksA in Zn homeostasis and stringent response.
Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crécy-Lagard V. Blaby-Haas CE, et al. Mol Microbiol. 2011 Feb;79(3):700-15. doi: 10.1111/j.1365-2958.2010.07475.x. Epub 2010 Dec 7. Mol Microbiol. 2011. PMID: 21255113 Free PMC article. - Transcriptional Regulation Contributes to Prioritized Detoxification of Hydrogen Peroxide over Nitric Oxide.
Adolfsen KJ, Chou WK, Brynildsen MP. Adolfsen KJ, et al. J Bacteriol. 2019 Jun 21;201(14):e00081-19. doi: 10.1128/JB.00081-19. Print 2019 Jul 15. J Bacteriol. 2019. PMID: 31061166 Free PMC article.
References
- Balandina, A., L. Claret, R. Hengge-Aronis, and J. Rouviere-Yaniv. 2001. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 39:1069-1079. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases