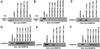Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases - PubMed (original) (raw)
Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases
Gun-Do Kim et al. EMBO J. 2002.
Abstract
Three different families of DNA (cytosine-5) methyltransferases, DNMT1, DUMT2, DNMT3a and DNMT3b, participate in establishing and maintaining genomic methylation patterns during mammalian development. These enzymes have a large N-terminal domain fused to a catalytic domain. The catalytic domain is homologous to prokaryotic (cytosine-5) methyltransferases and contains the catalytic PC dipeptide, while the N-terminus acts as a transcriptional repressor by recruiting several chromatin remodeling proteins. Here, we show that the human de novo enzymes hDNMT3a and hDNMT3b form complexes with the major maintenance enzyme hDNMT1. Antibodies against hDNMT1 pull down both the de novo enzymes. Furthermore, the N-termini of the enzymes are involved in protein-protein interactions. Immunocytochemical staining revealed mostly nuclear co-localization of the fusion proteins, with the exception of hDNMT3a, which is found either exclusively in cytoplasm or in both nucleus and cytoplasm. Pre-methylated substrate DNAs exhibited differential methylation by de novo and maintenance enzymes. In vivo co-expression of hDNMT1 and hDNMT3a or hDNMT3b leads to methylation spreading in the genome, suggesting co-operation between de novo and maintenance enzymes during DNA methylation.
Figures
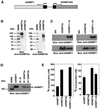
Fig. 1. Recombinant hDNMT expression and binding analysis in vivo. (A) Construct used for the expression of DNMTs in Sf9 cells. hDNMT1 expression was driven by the baculovirus p10 promoter. In the same construct, the polyhedrin (pPH) promoter expressed hDNMT3a or hDNMT3b, as indicated by the arrow. (B) Specificity of hDNMT3 antibodies. Proteins, hDNMT3a or hDNMT3b are indicated at the top, and the antibody used at the bottom, of each panel. Each lane had 0.4 µg of purified antigen. Molecular weight markers are shown on the left. (C) Western blot analysis of the expression of the hDNMTs in insect cell extracts. Extracts from Sf9 cells expressing hDNMT3a alone, hDNMT3b alone, or DNMT1 plus hDNMT3a or hDNMT3b, are indicated at the top, and the antibody used at the bottom, of each panel. The Sf9 lane is a control cell extract. (D) Co-immunoprecipitation with anti-DNMTs in Sf9 cells expressing recombinant enzyme(s). Antibodies used for immunoprecipitation are indicated at the top. Monoclonal anti-GFP was used as a control. Insect cell extracts expressing hDNMT1 and hDNMT3a were immunoprecipitated with anti-hDNMT3a, and extracts from cells expressing hDNMT1 and DNMT3b were immunoprecipitated with anti-hDNMT3b. Pre-immune serum is marked as PI. The arrow shows the relative position of hDNMT1, along with purified hDNMT1 as a positive control. (E) Maintenance methyltransferase activity of co-immunoprecipitates with de novo enzymes. Extracts from Sf9 cells expressing hDNMT1 and hDNMT3a (left), or hDNMT1 and hDNMT3b (right), were immunoprecipitated with anti-DNMTs antibodies, as indicated. The immunoprecipitated product was assayed for hDNMT1 activity using poly (dI–dC)·poly (dI–dC) substrate DNA and tritiated AdoMet.
Fig. 2. Expression profile and interactions between DNMTs in human cells. (A) Western blot analyses of DNMT1 (top), DNMT3a (middle) and DNMT3b (bottom) are shown. Nuclear extracts from different cell lines were used as indicated at the top, along with the positive control proteins hDNMT1, hDNMT3a and hDNMT3b. Molecular weight markers (in kDa) are in the extreme left and right lanes. Arrows indicate the relative locations of the endogenous DNMTs. Note that the same western blot gel was probed with three different antibodies for relative quantitation of the DNMTs. (B) Immunoprecipitation of hDNMTs in HEK-293 cell nuclear extracts. Antibodies used are indicated at the top. Control hDNMT1, hDNMT3a and hDNMT3b proteins were used as reference. Arrows on the right show the relative locations of the protein. Anti-hDNMT1, -hDNMT3a and -hDNMT3b antibodies were used for detection. Two additional arrows on the hDNMT3b western blot indicate the splice variants of hDNMT3b.
Fig. 3. Detailed mapping of the interacting regions of DNMTs. The GST–DNMT fusion peptides used for complex formation are indicated above each blot, with amino acid residue numbers shown in parentheses. Positive and negative controls are purified hDNMT1, hDNMT3a, hDNMT3b and GST protein, respectively. Molecular weight markers are in kDa. Antibodies used for the blots are indicated below each panel. (A) Immunoblot of bound hDNMT1 to various GST fusion fragments of hDNMT3a. (B) A similar immunoblot to that of (A) showing various GST fusion peptides of hDNMT3b bound to hDNMT1. (C) Identification of the hDNMT3a binding region of hDNMT1. (D) Finer mapping of the hDNMT1 binding region of hDNMT3a. (E) Immunoblot of bound hDNMT3b to various GST fusion fragments of hDNMT1. (F) Finer mapping of the hDNMT1 binding region of hDNMT3b. (G) hDNMT3a binding region of hDNMT3b. (H) hDNMT3b binding region of hDNMT3a. (I) Summary diagram of the various functional regions of DNMTs. Amino acid residues are indicated for various DNMTs. The relative locations of binding domains for other proteins are also indicated for reference. Proliferating cell nuclear antigen (PCNA), retinoblastoma gene product (Rb1), histone deacetylase (HDAC), DNA methyltransferase associated protein (DMAP) and methylated DNA-dependent allosteric activation domain (MDDAAD) are indicated. Another Rb binding region (Rb2) was recently discovered and mapped at the N-terminus of hDNMT1. Amino acid residues for protein–protein interaction are indicated for all hDNMTs.
Fig. 3. Detailed mapping of the interacting regions of DNMTs. The GST–DNMT fusion peptides used for complex formation are indicated above each blot, with amino acid residue numbers shown in parentheses. Positive and negative controls are purified hDNMT1, hDNMT3a, hDNMT3b and GST protein, respectively. Molecular weight markers are in kDa. Antibodies used for the blots are indicated below each panel. (A) Immunoblot of bound hDNMT1 to various GST fusion fragments of hDNMT3a. (B) A similar immunoblot to that of (A) showing various GST fusion peptides of hDNMT3b bound to hDNMT1. (C) Identification of the hDNMT3a binding region of hDNMT1. (D) Finer mapping of the hDNMT1 binding region of hDNMT3a. (E) Immunoblot of bound hDNMT3b to various GST fusion fragments of hDNMT1. (F) Finer mapping of the hDNMT1 binding region of hDNMT3b. (G) hDNMT3a binding region of hDNMT3b. (H) hDNMT3b binding region of hDNMT3a. (I) Summary diagram of the various functional regions of DNMTs. Amino acid residues are indicated for various DNMTs. The relative locations of binding domains for other proteins are also indicated for reference. Proliferating cell nuclear antigen (PCNA), retinoblastoma gene product (Rb1), histone deacetylase (HDAC), DNA methyltransferase associated protein (DMAP) and methylated DNA-dependent allosteric activation domain (MDDAAD) are indicated. Another Rb binding region (Rb2) was recently discovered and mapped at the N-terminus of hDNMT1. Amino acid residues for protein–protein interaction are indicated for all hDNMTs.
Fig. 4. Order of binding of hDNMTs. The GST–DNMT fusion peptides of hDNMT3a and hDNMT3b used for enzyme complex formation are indicated above each blot, with amino acid residue numbers shown in parentheses. Positive and negative controls are purified hDNMT1, hDNMT3a, hDNMT3b and GST protein, respectively. Molecular weight markers are in kDa. Antibodies used for the blots are indicated below each panel. (A) Immunoblot of bound hDNMT1 to various GST fusion fragments of hDNMT3a on GST beads. (B) A similar immunoblot showing various GST fusion peptides of hDNMT3b on GST beads bound to hDNMT1. (C) Identification of the full-length hDNMT3b binding to the hDNMT1–DNMT3a complex. (D) Identification of the full-length hDNMT3a binding to the hDNMT1–DNMT3b complex.
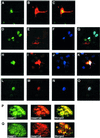
Fig. 5. Representative confocal laser scanning microscopy images of COS-7 cells transfected with pGFPDNMT446 (hDNMT1), pcDNAGST3A403 (hDNMT3a) and pcDNAHis3b515 (hDNMT3b), or a combination of these clones expressing fusion/tagged hDNMT proteins. hDNMT1 appears in green, hDNMT3a in red and hDNMT3b in blue. Merged images of hDNMT1 and hDNMT3a are greenish-yellow in color; those of hDNMT1 and hDNMT3b are whitish-blue. Merged images of hDNMT1, hDNMT3a and hDNMT3b are white. Transfection and localization of hDNMT1 and hDNMT3a. (A) GFP–hDNMT1, (B) GST–hDNMT3a and (C) merge of hDNMT1 and hDNMT3a. Note that hDNMT3a is both nuclear as well as cytoplasmic. Transfection of all three hDNMTs: (D) GFP–hDNMT1, (E) GST–hDNMT3a, (F) His–hDNMT3b and (G) merge of all the hDNMTs. Note that hDNMT3a is only cytoplasmic (E and G). Another set of transfections showing all three hDNMTs: (H) GFP–hDNMT1, (I) GST–hDNMT3a, (J) His–hDNMT3b and (K) merge of all the hDNMTs. Note that in this set DNMT3a is localized both in the nucleus as well as cytoplasm (I and K). All three hDNMTs show nuclear co-localization. In another set of transfections, all three hDNMTs exclusively localized in the nucleus: (L) GFP–hDNMT1, (M) GST–hDNMT3a, (N) His–hDNMT3b and (O) merge of all the hDNMT1. (P and Q) Nuclear co-localization of hDNMTs. Expressions of GFP–hDNMT1 in green and GST–hDNMT3a or His–hDNMT3b in red are shown. The merged expression pattern is in yellow or greenish-yellow.
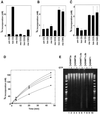
Fig. 6. Differential methylation of pre-methylated substrate DNA in vitro and methylation spreading in vivo. In vitro methylation using pre-methylated substrate with (A) hDNMT1, (B) hDNMT3a or (C) hDNMT3b. Equimolar concentrations of DNMTs were added to the reaction mix. Incorporation was measured after 30 min for hDNMT1 and 1 h for hDNMT3a or hDNMT3b. The experiment was conducted in duplicate and the average of two independent experiments is reported. (D) Co-operation for DNA methylation spreading in vitro. Reaction velocity curve of hDNMT1 and hDNMT3b (filled diamonds), hDNMT1, hDNMT3a and hDNMT3b (open squares), hDNMT1 and hDNMT3a (filled squares), hDNMT1 (filled circles), hDNMT3a (open circles), hDNMT3a and hDNMT3b (filled triangle), and hDNMT3b (open diamonds). Methylation by hDNMT3a, hDNMT3b and hDNMT3a, and hDNMT3b were similar and the data points are overlapping. The average values of two independent data points are plotted. The rates of methylation are shown in Table I. (E) Methylation spreading in vivo. Genomic DNAs from insect cells expressing different hDNMTs (indicated on top) were purified and digested with McrBC in the presence (+) or absence (–) of cofactor GTP. The digested DNA was analyzed on a 1.2% agarose gel. A 10 kbp to 100 bp ladder was used as a marker. um, unmethylated; hm, heminethylated; me, methylated.
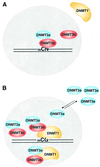
Fig. 7. Model for DNA methylation. (A) mCpN methylation by DNMT3a and DNMT3b. DNMT3s can catalyze mCG, mCA, mCT and mCC methylation in vivo, either individually or together. This type of methylation is predominant in the pre-implantation stage of mammalian development. A shorter DNMT1, lacking 118 amino acids, stays in the cytoplasm, except at the 8-cell stage of the embryo (Mertineit et al., 1998). Lack of this N-terminal region might lead to an unfavorable interaction between de novo and maintenance methyltransferases. (B) DNA methylation by DNMTs. Both DNMT1 and DNMT3b co-localize in the nucleus. DNMT3a may interact with either DNMT3b or the full-length DNMT1. Excess DNMT3a is escorted out to the cytoplasm, or cytoplasmic DNMT3a is not allowed into the nucleus. DNMT1 ensures CG methylation in the cell. The nuclear membrane is shown as a dotted circle.
Similar articles
- Hybrid mouse-prokaryotic DNA (cytosine-5) methyltransferases retain the specificity of the parental C-terminal domain.
Pradhan S, Roberts RJ. Pradhan S, et al. EMBO J. 2000 May 2;19(9):2103-14. doi: 10.1093/emboj/19.9.2103. EMBO J. 2000. PMID: 10790376 Free PMC article. - Allosteric activator domain of maintenance human DNA (cytosine-5) methyltransferase and its role in methylation spreading.
Pradhan S, Estève PO. Pradhan S, et al. Biochemistry. 2003 May 13;42(18):5321-32. doi: 10.1021/bi034160+. Biochemistry. 2003. PMID: 12731873 - Mammalian DNA methyltransferases show different subnuclear distributions.
Margot JB, Cardoso MC, Leonhardt H. Margot JB, et al. J Cell Biochem. 2001 Aug 21-Sep 5;83(3):373-9. doi: 10.1002/jcb.1236. J Cell Biochem. 2001. PMID: 11596106 - Molecular enzymology of mammalian DNA methyltransferases.
Jeltsch A. Jeltsch A. Curr Top Microbiol Immunol. 2006;301:203-25. doi: 10.1007/3-540-31390-7_7. Curr Top Microbiol Immunol. 2006. PMID: 16570849 Review. - [DNA methyltransferases: classification, functions and research progress].
Wang ZG, Wu JX. Wang ZG, et al. Yi Chuan. 2009 Sep;31(9):903-12. doi: 10.3724/sp.j.1005.2009.00903. Yi Chuan. 2009. PMID: 19819843 Review. Chinese.
Cited by
- Zinc Finger Readers of Methylated DNA.
Hudson NO, Buck-Koehntop BA. Hudson NO, et al. Molecules. 2018 Oct 7;23(10):2555. doi: 10.3390/molecules23102555. Molecules. 2018. PMID: 30301273 Free PMC article. Review. - A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis.
Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. Coward WR, et al. FASEB J. 2014 Jul;28(7):3183-96. doi: 10.1096/fj.13-241760. Epub 2014 Mar 20. FASEB J. 2014. PMID: 24652950 Free PMC article. - The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes.
Kundakovic M, Chen Y, Guidotti A, Grayson DR. Kundakovic M, et al. Mol Pharmacol. 2009 Feb;75(2):342-54. doi: 10.1124/mol.108.051763. Epub 2008 Nov 24. Mol Pharmacol. 2009. PMID: 19029285 Free PMC article. - The epigenetics of adult (somatic) stem cells.
Eilertsen KJ, Floyd Z, Gimble JM. Eilertsen KJ, et al. Crit Rev Eukaryot Gene Expr. 2008;18(3):189-206. doi: 10.1615/critreveukargeneexpr.v18.i3.10. Crit Rev Eukaryot Gene Expr. 2008. PMID: 18540823 Free PMC article. Review. - Repeated PM2.5 exposure inhibits BEAS-2B cell P53 expression through ROS-Akt-DNMT3B pathway-mediated promoter hypermethylation.
Zhou W, Tian D, He J, Wang Y, Zhang L, Cui L, Jia L, Zhang L, Li L, Shu Y, Yu S, Zhao J, Yuan X, Peng S. Zhou W, et al. Oncotarget. 2016 Apr 12;7(15):20691-703. doi: 10.18632/oncotarget.7842. Oncotarget. 2016. PMID: 26942697 Free PMC article.
References
- Bacolla A., Pradhan,S., Roberts,R.J. and Wells,R.D. (1999) Recombinant human DNA (cytosine-5) methyltransferase II. Steady-state kinetics reveal allosteric activation by methylated DNA. J. Biol. Chem., 274, 33011–33019. - PubMed
- Bacolla A., Pradhan,S., Larson,J.E., Roberts,R.J. and Wells,R.D. (2001) Recombinant human DNA (cytosine-5) methyltransferase II. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J. Biol. Chem., 276, 18605–18608. - PubMed
- Baylin S.B., Herman,J.G., Graff,J.R., Vertino,P.M. and Issa,J.P. (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res., 72, 141–196. - PubMed
- Bonfils C., Beaulieu,N., Chan,E., Cotton-Montpetit,J. and MacLeod,A.R. (2000) Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem., 275, 10754–10760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases