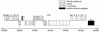Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates - PubMed (original) (raw)
Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates
Diane E Taylor et al. J Bacteriol. 2002 Sep.
Abstract
Strains of Escherichia coli causing enterohemorrhagic colitis belonging to the O157:H7 lineage are reported to be highly related. Fifteen strains of E. coli O157:H7 and 1 strain of E. coli O46:H(-) (nonflagellated) were examined for the presence of potassium tellurite resistance (Te(r)). Te(r) genes comprising terABCDEF were shown previously to be part of a pathogenicity island also containing integrase, phage, and urease genes. PCR analysis, both conventional and light cycler based, demonstrated that about one-half of the Te(r) E. coli O157:H7 strains (6 of 15), including the Sakai strain, which has been sequenced, carried a single copy of the Te(r) genes. Five of the strains, including EDL933, which has also been sequenced, contained two copies. Three other O157:H7 strains and the O46:H(-) strain did not contain the Te(r) genes. In strains containing two copies, the Te(r) genes were associated with the serW and serX tRNA genes. Five O157:H7 strains resembled the O157 Sakai strain whose sequence contained one copy, close to serX, whereas in one isolate the single copy was associated with serW. There was no correlation between Te(r) and the ability to produce Shiga toxin ST1 or ST2. The Te(r) MIC for most strains, containing either one or two copies, was 1,024 micro g/ml, although for a few the MIC was intermediate, 64 to 128 micro g/ml, which could be increased to 512 micro g/ml by pregrowth of strains in subinhibitory concentrations of potassium tellurite. Reverse transcriptase PCR analysis confirmed that in most strains Te(r) was constitutive but that in the rest it was inducible and involved induction of terB and terC genes. Only the terB, -C, -D, and -E genes are required for Te(r). The considerable degree of homology between the ter genes on IncH12 plasmid R478, which originated in Serratia marcescens, and pTE53, from an E. coli clinical isolate, suggests that the pathogenicity island was acquired from a plasmid. This work demonstrates diversity among E. coli O157:H7 isolates, at least as far as the presence of Te(r) genes is concerned.
Figures
FIG. 1.
Map of OI 43 and 48 from E. coli O157: H7 strain EDL933 (14). Genes of interest encode urease, a ribosomal protein, tellurite resistance, an insertion sequence, and adhesin (iha). A putative ribosomal protein gene (unlabeled) downstream of the urease operon is 96% identical to E. coli K-12 ORF ygkM. The overall size of the islands is 87,562 bp. The region from bp 20500 to 50000 is shown.
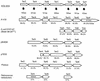
FIG. 2.
Comparison of Ter determinants from various sources. Deduced ORFs are shown, with the sizes in amino acids of the products of ORFs from E. coli O157:H7 EDL933 (14) below the filled arrows. ORFs from Serratia marcescens plasmid R478 (24) are most closely related. The percent identity of each ORF to its corresponding ORF in EDL933 is shown below. Those labeled TlpD and TlrC, -B, and -A from E. coli O157: H7 strain 86-24NalR were given this nomenclature by Tarr et al. (19). Plasmid pMJ606 contains the cloned Ter determinants from pMER610 originally isolated from an Alcaligenes sp. (7). Plasmid pTE53 originated in a clinical isolate of E. coli (3). The last two sequences are from Proteus sp. and D. radiodurans genome sequence databases.
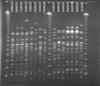
FIG. 3.
PFGE of E. coli O157:H7 DNA digested with _Xba_I. The strain numbers are shown at the top of the gel. Lambda phage size markers were run at each end of the gel and in the central well.
Similar articles
- Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food.
Orth D, Grif K, Dierich MP, Würzner R. Orth D, et al. Res Microbiol. 2007 Mar;158(2):105-11. doi: 10.1016/j.resmic.2006.10.007. Epub 2006 Dec 20. Res Microbiol. 2007. PMID: 17317110 - Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates.
Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. Bielaszewska M, et al. J Clin Microbiol. 2005 Jan;43(1):452-4. doi: 10.1128/JCM.43.1.452-454.2005. J Clin Microbiol. 2005. PMID: 15635013 Free PMC article. - [Variability in tellurite resistance and the ter gene cluster among Escherichia coli O157 isolated in food from 2005 to 2007].
Bai L, Dong Y, Guo Y. Bai L, et al. Wei Sheng Yan Jiu. 2011 May;40(3):287-9. Wei Sheng Yan Jiu. 2011. PMID: 21695895 Chinese. - Increased resistance against tellurite is conferred by a mutation in the promoter region of uncommon tellurite resistance gene tehB in the _ter_-negative Shiga toxin-producing Escherichia coli O157:H7.
Matsumoto Y, Lee K, Akasaka R, Honjo H, Koizumi M, Sato T, Kubomura A, Ishijima N, Akeda Y, Ohnishi M, Iyoda S. Matsumoto Y, et al. Appl Environ Microbiol. 2024 Jun 18;90(6):e0228323. doi: 10.1128/aem.02283-23. Epub 2024 May 17. Appl Environ Microbiol. 2024. PMID: 38757978 - A homolog of the O157 urease-encoding O island 48 is present in porcine O149:H10 enterotoxigenic Escherichia coli.
Parreira VR, Liao JH, Kim SH, Gyles CL. Parreira VR, et al. Vet Res. 2008 May-Jun;39(3):38. doi: 10.1051/vetres:2008015. Epub 2008 Mar 4. Vet Res. 2008. PMID: 18316019
Cited by
- The tellurite resistance gene cluster of pathogenic bacteria and its effect on oxidative stress response.
Vávrová S, Grones J, Šoltys K, Celec P, Turňa J. Vávrová S, et al. Folia Microbiol (Praha). 2024 Apr;69(2):433-444. doi: 10.1007/s12223-024-01133-8. Epub 2024 Jan 23. Folia Microbiol (Praha). 2024. PMID: 38261148 Free PMC article. - Pathogenicity assessment of Shiga toxin-producing Escherichia coli strains isolated from wild birds in a major agricultural region in California.
Carter MQ, Quiñones B, Laniohan N, Carychao D, Pham A, He X, Cooley M. Carter MQ, et al. Front Microbiol. 2023 Sep 26;14:1214081. doi: 10.3389/fmicb.2023.1214081. eCollection 2023. Front Microbiol. 2023. PMID: 37822735 Free PMC article. - Global population structure, genomic diversity and carbohydrate fermentation characteristics of clonal complex 119 (CC119), an understudied Shiga toxin-producing E. coli (STEC) lineage including O165:H25 and O172:H25.
Nakamura K, Seto K, Lee K, Ooka T, Gotoh Y, Taniguchi I, Ogura Y, Mainil JG, Piérard D, Harada T, Etoh Y, Ueda S, Hamasaki M, Isobe J, Kimata K, Narimatsu H, Yatsuyanagi J, Ohnishi M, Iyoda S, Hayashi T. Nakamura K, et al. Microb Genom. 2023 Mar;9(3):mgen000959. doi: 10.1099/mgen.0.000959. Microb Genom. 2023. PMID: 36951916 Free PMC article. - Comparative genomic and phenotypic analyses of the virulence potential in Shiga toxin-producing Escherichia coli O121:H7 and O121:H10.
Carter MQ, Laniohan N, Pham A, Quiñones B. Carter MQ, et al. Front Cell Infect Microbiol. 2022 Nov 24;12:1043726. doi: 10.3389/fcimb.2022.1043726. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36506028 Free PMC article. - Enhancing tellurite and selenite bioconversions by overexpressing a methyltransferase from Aromatoleum sp. CIB.
Alonso-Fernandes E, Fernández-Llamosas H, Cano I, Serrano-Pelejero C, Castro L, Díaz E, Carmona M. Alonso-Fernandes E, et al. Microb Biotechnol. 2023 May;16(5):915-930. doi: 10.1111/1751-7915.14162. Epub 2022 Nov 10. Microb Biotechnol. 2023. PMID: 36366868 Free PMC article.
References
- Alonso, G., C. Gomez, C. Gonzalez, and V. R. Lemoine. 2000. On the mechanism of resistance to channel-forming colicins (PacB) and tellurite, encoded by plasmid Mip233 (IncHI3). FEMS Microbiol. Lett. 192:257-261. - PubMed
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. Goeden, D. Rose, B. Mau, and Y. Shao. 1997. The complete sequence of Escherichia coli K-12. Science 277:1453-1462. - PubMed
- Burian, J., N. Tu, L. Klucar, L. Guller, G. Lloyd-Jones, S. Stuchlik, P. Fejdi, P. Siekel, and J. Turna. 1998. In vivo and in vitro cloning and phenotype characterization of tellurite resistance determinant conferred by plasmid pTE53 of a clinical isolate of Escherichia coli. Folia Microbiol. 43:589-599. - PubMed
- Chang, N., Q. Jiang, and D. E. Taylor. 1997. Construction of a genomic map of H. pylori by pulsed-field gel electrophoresis (PFGE), p. 165-176. In C. L. Clayton and H. L. T. Mobley (ed.), Methods in molecular medicine, Helicobacter pylori protocols. Humana Press Inc., Totowa, N.J. - PubMed
- Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases