MLL-ENL cooperates with SCF to transform primary avian multipotent cells - PubMed (original) (raw)
MLL-ENL cooperates with SCF to transform primary avian multipotent cells
Cathleen E Schulte et al. EMBO J. 2002.
Abstract
The MLL gene is targeted by chromosomal translocations, which give rise to heterologous MLL fusion proteins and are associated with distinct types of acute lymphoid and myeloid leukaemia. To determine how MLL fusion proteins alter the proliferation and/or differentiation of primary haematopoietic progenitors, we introduced the MLL-AF9 and MLL-ENL fusion proteins into primary chicken bone marrow cells. Both fusion proteins caused the sustained outgrowth of immature haematopoietic cells, which was strictly dependent on stem cell factor (SCF). The renewing cells have a long in vitro lifespan exceeding the Hayflick limit of avian cells. Analysis of clonal cultures identified the renewing cells as immature, multipotent progenitors, expressing erythroid, myeloid, lymphoid and stem cell surface markers. Employing a two-step commitment/differentiation protocol involving the controlled withdrawal of SCF, the MLL-ENL-transformed progenitors could be induced to terminal erythroid or myeloid differentiation. Finally, in cooperation with the weakly leukaemogenic receptor tyrosine kinase v-Sea, the MLL-ENL fusion protein gave rise to multilineage leukaemia in chicks, suggesting that other activated, receptor tyrosine kinases can substitute for ligand-activated c-Kit in vivo.
Figures
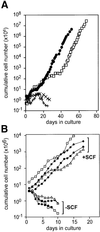
Fig. 1. Growth of MLL–AF9- and MLL–ENL-infected cultures. (A) MLL–AF9- (solid circles) and MLL–ENL (open squares)-infected mass cultures, as well as control cultures infected with empty vector (+ and ×), were cultivated in multipotent cell medium and cumulative cell numbers determined daily by counting in an electronic counter (see Materials and methods). (B) Four clones verified for single MLL–ENL integration sites: D8 (triangles), D11 (diamonds), B8 (circles) and IIC10 (squares) were cultured in multipotent cell medium containing (+SCF) or lacking (–SCF) SCF. Cumulative cell numbers were determined as above.
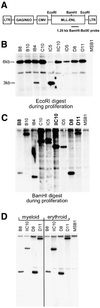
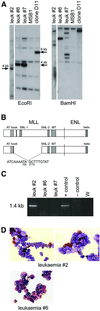
Fig. 2. Molecular analysis of MLL–ENL-infected cultures. (A) Schematic representation of the CRNCM-MLL–ENL retroviral vector used in this study. (B) The presence of full-length MLL–ENL cDNA in nine multipotential methocel clones was detected by Southern blot analysis, digesting genomic DNA with _Eco_RI and hybridizing with a human MLL–ENL probe (1.25 kb _Bam_HI–_Bst_XI fragment, see A). (C and D) Monoclonal origin of MLL–ENL-infected clonal cultures. To identify clones containing single retroviral integration sites, genomic DNA from nine clones grown in multipotential medium (C) and from four clones after terminal differentiation (D) into myeloid (left panel) or erythroid cells (right panel; see Figure 4 and Materials and methods) was subjected to Southern blot analysis after digestion with _Bam_HI and hybridization with the same human MLL–ENL probe as in (B). Genomic DNA from the chicken T-cell line MSB1 was used as control for cross-reactive endogenous chicken MLL.
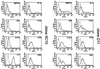
Fig. 3. MLL–ENL-infected cultures express multilineage cell surface markers. Cell surface marker profiles for MLL–ENL clones IIC10 and D11 as determined by FACS analysis. For a detailed description of antigens detected by the panel of antibodies used, see Results.
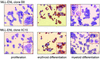
Fig. 4. MLL–ENL-transformed multipotent clones can be induced to differentiate along erythroid and myeloid lineages. Cells from MLL–ENL clones B8 (top panels) and IIC10 (bottom panels) were cytocentrifuged on to slides after cultivation in multipotent cell medium (left panels) or after induction of terminal erythroid (middle panels) or myeloid differentiation (right panels), employing a two-step commitment/differentiation protocol (Beug et al., 1995a; Materials and methods). Photographs of representative cytospins are shown after staining with acid benzidine (haemoglobin, yellow-brown) plus Wright–Giemsa (Beug et al., 1995b). Insets: cells at higher magnification.
Fig. 5. Molecular and phenotypic analysis of MLL–ENL/ts-v-Sea- induced leukaemias. (A) Southern blot of genomic DNA extracted from bone marrow of leukaemic chickens. Digestion with _Eco_RI reveals a truncated MLL–ENL cDNA of ∼4 kb in all leukaemias analysed (see legend to Figure 2). As a control, the MLL–ENL clone D11 containing the 6 kb full-length MLL–ENL cDNA and the chicken T-cell line MSB1 are shown. (B) Schematic representation of _MLL–ENL_-specific sequences detected in leukaemias 2 and 6, revealing an in-frame deletion. See Results and Supplementary data for a detailed description of the respective nested PCR analyses. (C) Analysis of ts-v-Sea in leukaemias 2, 6 and 7 by PCR analysis generated the expected 1.4 kb product, encompassing the S13 viral env sequence plus the transmembrane and complete tyrosine kinase domains of v-Sea. Positive control, _in vitro_-transformed ts-v-Sea/MLL–ENL clone; negative control, MLL–ENL clone D8; W, water control. (D) Cultured cells from leukaemia 2 (two fields, top) and leukaemia 6 (bottom) are shown after cytocentrifugation on to slides and staining with acid benzidine and Wright–Giemsa.
Fig. 6. ts-v-Sea can substitute for SCF-induced c-Kit signalling required for leukaemic transformation by MLL–ENL. _In vitro_-transformed MLL–ENL/ts-v-Sea clones C7, D6 and G8 were cultivated in S13 medium without additions at 37°C (triangles) or at 42°C in the same medium plus 300 µM of the glycosylation inhibitor castanospermine, in the presence (squares) or absence (circles) of SCF. Cumulative cell numbers were determined daily using an electronic cell counter.
Similar articles
- Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis.
Drynan LF, Pannell R, Forster A, Chan NM, Cano F, Daser A, Rabbitts TH. Drynan LF, et al. EMBO J. 2005 Sep 7;24(17):3136-46. doi: 10.1038/sj.emboj.7600760. Epub 2005 Aug 11. EMBO J. 2005. PMID: 16096649 Free PMC article. - Leukaemia lineage specification caused by cell-specific Mll-Enl translocations.
Cano F, Drynan LF, Pannell R, Rabbitts TH. Cano F, et al. Oncogene. 2008 Mar 20;27(13):1945-50. doi: 10.1038/sj.onc.1210818. Epub 2007 Oct 1. Oncogene. 2008. PMID: 17906700 - The oncoprotein MLL-ENL disturbs hematopoietic lineage determination and transforms a biphenotypic lymphoid/myeloid cell.
Zeisig BB, García-Cuéllar MP, Winkler TH, Slany RK. Zeisig BB, et al. Oncogene. 2003 Mar 20;22(11):1629-37. doi: 10.1038/sj.onc.1206104. Oncogene. 2003. PMID: 12642866 - Learning from mouse models of MLL fusion gene-driven acute leukemia.
Schwaller J. Schwaller J. Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194550. doi: 10.1016/j.bbagrm.2020.194550. Epub 2020 Apr 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32320749 Review. - MLL translocations, histone modifications and leukaemia stem-cell development.
Krivtsov AV, Armstrong SA. Krivtsov AV, et al. Nat Rev Cancer. 2007 Nov;7(11):823-33. doi: 10.1038/nrc2253. Nat Rev Cancer. 2007. PMID: 17957188 Review.
Cited by
- Molecular pathogenesis of MLL-associated leukemias.
Eguchi M, Eguchi-Ishimae M, Greaves M. Eguchi M, et al. Int J Hematol. 2005 Jul;82(1):9-20. doi: 10.1532/IJH97.05042. Int J Hematol. 2005. PMID: 16105754 Review. - Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis.
Ono R, Nakajima H, Ozaki K, Kumagai H, Kawashima T, Taki T, Kitamura T, Hayashi Y, Nosaka T. Ono R, et al. J Clin Invest. 2005 Apr;115(4):919-29. doi: 10.1172/JCI22725. Epub 2005 Mar 10. J Clin Invest. 2005. PMID: 15761502 Free PMC article. - Erythroid progenitor renewal versus differentiation: genetic evidence for cell autonomous, essential functions of EpoR, Stat5 and the GR.
Dolznig H, Grebien F, Deiner EM, Stangl K, Kolbus A, Habermann B, Kerenyi MA, Kieslinger M, Moriggl R, Beug H, Müllner EW. Dolznig H, et al. Oncogene. 2006 May 11;25(20):2890-900. doi: 10.1038/sj.onc.1209308. Oncogene. 2006. PMID: 16407844 Free PMC article. - Translation initiation factor 4E inhibits differentiation of erythroid progenitors.
Blázquez-Domingo M, Grech G, von Lindern M. Blázquez-Domingo M, et al. Mol Cell Biol. 2005 Oct;25(19):8496-506. doi: 10.1128/MCB.25.19.8496-8506.2005. Mol Cell Biol. 2005. PMID: 16166632 Free PMC article. - Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors.
Cozzio A, Passegué E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Cozzio A, et al. Genes Dev. 2003 Dec 15;17(24):3029-35. doi: 10.1101/gad.1143403. Genes Dev. 2003. PMID: 14701873 Free PMC article.
References
- Armstrong S.A. et al. (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet., 30, 41–47. - PubMed
- Ashman L.K., Ferrao,P., Cole,S.R. and Cambareri,A.C. (2000) Effects of mutant c-Kit in early myeloid cells. Leuk. Lymphoma, 37, 233–243. - PubMed
- Ayton P.M. and Cleary,M.L. (2001) Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene, 20, 5695–5707. - PubMed
- Bauer A., Gandrillon,O., Samarut,J. and Beug,H. (2001) Nuclear receptors in hematopoietic development: cooperation with growth factor receptors in regulation of proliferation and differentiation. In Zon,L. (ed.), Hematopoiesis: A Developmental Approach. Oxford University Press, Oxford, pp. 368–390.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical