Minimal introns are not "junk" - PubMed (original) (raw)
Minimal introns are not "junk"
Jun Yu et al. Genome Res. 2002 Aug.
Abstract
Intron-size distributions for most multicellular (and some unicellular) eukaryotes have a sharp peak at their "minimal intron" size. Across the human population, these minimal introns exhibit an abundance of insertion-deletion polymorphisms, the effect of which is to maintain their optimal size. We argue that minimal introns affect function by enhancing the rate at which mRNA is exported from the cell nucleus.
Figures
Figure 1
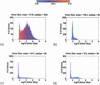
Intron size for Homo sapiens (a), Arabidopis thaliana (b), Drosophila melanogaster (c), and Caenorhabditis elegans (d). There is always a species-specific minimum intron size, at which a significant fraction of the introns tend to cluster. This “spike” in the distribution is centered around the mean (±SD) intron sizes of 92±14, 89±12, 61±10, and 48±9 bp, respectively. For larger introns, the size distribution is highly species-specific. In the extreme case, H. sapiens, there is a broad “hump” attributable to transposon insertions inside the introns. Color indicates GC content: Red is GC-rich; blue, AT-rich.
Figure 2
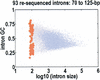
Resequenced introns, shown against the joint distribution for intron size and GC content. Introns with a detectible transposon are colored blue; introns with no detectible transposons, gray. The 93 introns that we resequenced are red. They are selected to sample the full range of GC contents, and their mean size is 94±14 bp.
Figure 3
Insertion-deletion (indel) direction, relative to the major allele, as a function of intron size. For the 10 rare indels of minor allele frequency f < 0.06 (blue), the resultant size changes drive the introns back to their optimal size of 92 bp. The two common indels with f > 0.35 (red) are the only exceptions to this rule, presumably because they arose from a different population dynamics.
Similar articles
- Mystery of intron gain.
Fedorov A, Roy S, Fedorova L, Gilbert W. Fedorov A, et al. Genome Res. 2003 Oct;13(10):2236-41. doi: 10.1101/gr.1029803. Epub 2003 Sep 15. Genome Res. 2003. PMID: 12975308 Free PMC article. - Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila.
Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A. Boudet N, et al. Genome Res. 2001 Dec;11(12):2101-14. doi: 10.1101/gr.200801. Genome Res. 2001. PMID: 11731501 Free PMC article. - Is "junk" DNA mostly intron DNA?
Wong GK, Passey DA, Huang Y, Yang Z, Yu J. Wong GK, et al. Genome Res. 2000 Nov;10(11):1672-8. doi: 10.1101/gr.148900. Genome Res. 2000. PMID: 11076852 Free PMC article. - [Post-splicing intron turnover].
Kataoka N. Kataoka N. Tanpakushitsu Kakusan Koso. 2006 Nov;51(14 Suppl):2210-2. Tanpakushitsu Kakusan Koso. 2006. PMID: 17471940 Review. Japanese. No abstract available. - Stable Intronic Sequence RNAs Engage in Feedback Loops.
Pek JW. Pek JW. Trends Genet. 2018 May;34(5):330-332. doi: 10.1016/j.tig.2018.01.006. Epub 2018 Jan 31. Trends Genet. 2018. PMID: 29397203 Review.
Cited by
- Pluripotency-Associated microRNAs in Early Vertebrate Embryos and Stem Cells.
Maraghechi P, Aponte MTS, Ecker A, Lázár B, Tóth R, Szabadi NT, Gócza E. Maraghechi P, et al. Genes (Basel). 2023 Jul 12;14(7):1434. doi: 10.3390/genes14071434. Genes (Basel). 2023. PMID: 37510338 Free PMC article. Review. - Intron size minimisation in teleosts.
Jakt LM, Dubin A, Johansen SD. Jakt LM, et al. BMC Genomics. 2022 Sep 1;23(1):628. doi: 10.1186/s12864-022-08760-w. BMC Genomics. 2022. PMID: 36050638 Free PMC article. - Evolution of the Early Spliceosomal Complex-From Constitutive to Regulated Splicing.
Borao S, Ayté J, Hümmer S. Borao S, et al. Int J Mol Sci. 2021 Nov 18;22(22):12444. doi: 10.3390/ijms222212444. Int J Mol Sci. 2021. PMID: 34830325 Free PMC article. Review. - SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns.
Fukumura K, Yoshimoto R, Sperotto L, Kang HS, Hirose T, Inoue K, Sattler M, Mayeda A. Fukumura K, et al. Nat Commun. 2021 Aug 13;12(1):4910. doi: 10.1038/s41467-021-24879-y. Nat Commun. 2021. PMID: 34389706 Free PMC article. - Expression of a human cDNA in moss results in spliced mRNAs and fragmentary protein isoforms.
Top O, Milferstaedt SWL, van Gessel N, Hoernstein SNW, Özdemir B, Decker EL, Reski R. Top O, et al. Commun Biol. 2021 Aug 12;4(1):964. doi: 10.1038/s42003-021-02486-3. Commun Biol. 2021. PMID: 34385580 Free PMC article.
References
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. - PubMed
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. - PubMed
- Blencowe BJ. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. - PubMed
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases