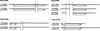Inferring alternative splicing patterns in mouse from a full-length cDNA library and microarray data - PubMed (original) (raw)
Inferring alternative splicing patterns in mouse from a full-length cDNA library and microarray data
Hiromi Kochiwa et al. Genome Res. 2002 Aug.
Abstract
Although many studies on alternative splicing of specific genes have been reported in the literature, the general mechanism that regulates alternative splicing has not been clearly understood. In this study, we systematically aligned each pair of the 21,076 cDNA sequences of Mus musculus, searched for putative alternative splicing patterns, and constructed a list of potential alternative splicing sites. Two cDNAs are suspected to be alternatively spliced and originating from a common gene if they share most of their region with a high degree of sequence homology, but parts of the sequences are very distinctive or deleted in either cDNA. The list contains the following information: (1) tissue, (2) developmental stage, (3) sequences around splice sites, (4) the length of each gapped region, and (5) other comments. The list is available at http://www.bioinfo.sfc.keio.ac.jp/intron. Our results have predicted a number of unreported alternatively spliced genes, some of which are expressed only in a specific tissue or at a specific developmental stage.
Figures
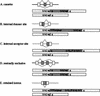
Figure 1
Patterns of alternative splicing. Nucleotide sequences are consensus sequences around the splicing sites (Mount 1982; Padgett et al. 1986).
Figure 2
An example of spliced and unspliced regions. Spliced has a gapped region.
Figure 3
Mutually exclusive splicing of the CHIP gene (Ballinger et al. 1999).
Figure 4
Examples of more complicated alternative splicing patterns in which three cDNAs were potentially produced in different forms from a single gene. Cluster 8: homologs to human PR domain zinc finger protein 5 (Deng et al., unpubl.). Cluster 45: homologs to human mitochondrial carrier homolog 2 (Jang et al., unpubl.). Cluster 63: homologs to human HSPC204 protein (Zhang et al. 2000). Cluster 74: homologs to human HSPC223 protein (Ye et al., unpubl.). Cluster 85: homologs to human heterogeneous nuclear ribonucleoprotein C (Nakagawa et al. 1986). Clusters 3022, 3058, and 3110: no homology found (hypothetical protein). Splice variant of Cluster 3058, no homology found (unclassifiable). Cluster 3147: homologs to D. melanogaster brain cDNA clone NMCB-2386 (Osada et al., unpubl.). Cluster 3148: homologs to bisphosphate 3′-nucleotidase (Spiegelberg et al. 1999).
Figure 5
These clusters each have a prominent splicing pattern in specific tissues or at distinct developmental stages. Cluster 2204: homologs to prolactin-like-peptide (Ishibashi and Imai 1999). Cluster 3082: homologs to human HSPC011 and 28S ribosomal protein S17, mitochondrial precursor (Gantt and Thompson 1990). Cluster 3138: homologs to TIA-1 cytotoxic granule-associated RNA-binding protein-like 1 (Lowin et al. 1996). Cluster 3148: homologs to bisphosphate 3′-nucleotidase (Spiegelberg et al. 1999).
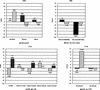
Figure 6
The horizontal axis is the tissue in which the gene expression was observed. The vertical axis is the level of gene expression as a score of signal intensity between cDNAs (log).
Similar articles
- Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes.
Itoh H, Washio T, Tomita M. Itoh H, et al. RNA. 2004 Jul;10(7):1005-18. doi: 10.1261/rna.5221604. RNA. 2004. PMID: 15208437 Free PMC article. - Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants.
Wistow G, Bernstein SL, Wyatt MK, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K. Wistow G, et al. Mol Vis. 2002 Jun 15;8:171-84. Mol Vis. 2002. PMID: 12107413 - Efficient filtering methods for clustering cDNAs with spliced sequence alignment.
Shibuya T, Kashima H, Konagaya A. Shibuya T, et al. Bioinformatics. 2004 Jan 1;20(1):29-39. doi: 10.1093/bioinformatics/btg367. Bioinformatics. 2004. PMID: 14693805 - Bioinformatics analysis of alternative splicing.
Lee C, Wang Q. Lee C, et al. Brief Bioinform. 2005 Mar;6(1):23-33. doi: 10.1093/bib/6.1.23. Brief Bioinform. 2005. PMID: 15826354 Review. - Transcriptome analyses of human genes and applications for proteome analyses.
Suzuki Y, Sugano S. Suzuki Y, et al. Curr Protein Pept Sci. 2006 Apr;7(2):147-63. doi: 10.2174/138920306776359795. Curr Protein Pept Sci. 2006. PMID: 16611140 Review.
Cited by
- Selecting for functional alternative splices in ESTs.
Kan Z, States D, Gish W. Kan Z, et al. Genome Res. 2002 Dec;12(12):1837-45. doi: 10.1101/gr.764102. Genome Res. 2002. PMID: 12466287 Free PMC article. - Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data.
Le K, Mitsouras K, Roy M, Wang Q, Xu Q, Nelson SF, Lee C. Le K, et al. Nucleic Acids Res. 2004 Dec 14;32(22):e180. doi: 10.1093/nar/gnh173. Nucleic Acids Res. 2004. PMID: 15598820 Free PMC article. - Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes.
Itoh H, Washio T, Tomita M. Itoh H, et al. RNA. 2004 Jul;10(7):1005-18. doi: 10.1261/rna.5221604. RNA. 2004. PMID: 15208437 Free PMC article. - Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs.
Takeda J, Suzuki Y, Nakao M, Barrero RA, Koyanagi KO, Jin L, Motono C, Hata H, Isogai T, Nagai K, Otsuki T, Kuryshev V, Shionyu M, Yura K, Go M, Thierry-Mieg J, Thierry-Mieg D, Wiemann S, Nomura N, Sugano S, Gojobori T, Imanishi T. Takeda J, et al. Nucleic Acids Res. 2006;34(14):3917-28. doi: 10.1093/nar/gkl507. Epub 2006 Aug 12. Nucleic Acids Res. 2006. PMID: 16914452 Free PMC article. - Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing.
Castle J, Garrett-Engele P, Armour CD, Duenwald SJ, Loerch PM, Meyer MR, Schadt EE, Stoughton R, Parrish ML, Shoemaker DD, Johnson JM. Castle J, et al. Genome Biol. 2003;4(10):R66. doi: 10.1186/gb-2003-4-10-r66. Epub 2003 Sep 19. Genome Biol. 2003. PMID: 14519201 Free PMC article.
References
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources