Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster - PubMed (original) (raw)
Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster
James T Warren et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Five different enzymatic activities, catalyzed by both microsomal and mitochondrial cytochrome P450 monooxygenases (CYPs), are strongly implicated in the biosynthesis of ecdysone (E) from cholesterol. However, none of these enzymes have been characterized completely. The present data show that the wild-type genes of two members of the Halloween family of embryonic lethals, disembodied (dib) and shadow (sad), code for mitochondrial cytochromes P450 that mediate the last two hydroxylation reactions in the ecdysteroidogenic pathway in Drosophila, namely the C22- and C2-hydroxylases. When sad (CYP315A1) is transfected into Drosophila S2 cells, the cells metabolize 2-deoxyecdysone (2dE) to E and the [3H]ketotriol (2,22-dideoxyecdysone) to 22-deoxyecdysone. In contrast, dib (CYP302A1) is responsible for the conversion of the [3H]ketotriol to [3H]2dE. When cells are transfected with both dib and sad, they metabolize the [3H]ketotriol to [3H]E in high yield. The expression of sad and dib is concentrated within the individual segments of the developing epidermis when there is a surge of ecdysteroid midway through embryogenesis. This result occurs before the ring gland has developed and suggests that the embryonic epidermis is a site of ecdysteroid biosynthesis. This pattern then diminishes, and, during late embryogenesis, expression of both genes is concentrated in the prothoracic gland cells of the developing ring gland. Expression of dib and sad continues to be localized in this endocrine compartment during larval development, being maximal in both the late second and third instar larvae, about the time of the premolt peaks in the ecdysteroid titer.
Figures
Fig 1.
Theoretical scheme of ecdysone biosynthesis (2). Dietary cholesterol or phytosterols serve as the ultimate precursors for the synthesis of 20E, because insects cannot synthesize sterols from simple carbon molecules. The intermediate resulting from the subsequent oxidation of 7dC has not been identified unequivocally, but a number of in vivo and in vitro studies suggest that: (i) it is the ketodiol (2,22,25dE) or its 3-dehydro derivative; (ii) these complex reactions are catalyzed by one or more CYP enzymes present within the mitochondria; and (iii) the delivery of 7dC from the endoplasmic reticulum to these enzymes may be rate-limiting for the production of E. The sequential hydroxylations of the ketodiol at carbon 25 (to the ketotriol), carbon 22 (to 2dE by dib), carbon 2 (to E by sad), carbon 20 (activation to 20E), and carbon 26 (inactivation to 20,26E) are catalyzed by CYP enzymes present in the microsomal and/or mitochondrial compartments (2, 5).
Fig 2.
(A) In situ expression pattern of sad. (a) Cellular blastoderm stage embryo; (b) germ band extension stage 8, segmental staining appears in the epidermal cells; (c) stage 10–11 embryo showing segment polarity type expression; (d) ring gland expression in a stage 17 embryo; (e) expression in the prothoracic gland cells of the ring gland of a third instar larva; (f) stage 10 ovary. (B) Cycling of dib and sad expression between the second (L2) and third (L3) instar. In situ hybridization pattern of dib (a_–_c) and sad (d_–_f) within the prothoracic gland cells of the ring gland. (a and d) Late L2 larval brain-ring gland complex; (b and e) early L3 larval brain-ring gland complex; (c and f) late L3 larval brain-ring gland complex. Rg, Ring gland; br, brain; vg, ventral ganglion.
Fig 3.
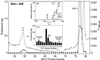
RP-HPLC/TLC/RIA/MS analysis after sad- or GFP-transfected S2 cell incubation (8 h) with the 2dE substrate (30–100% methanol gradient). Ecdysteroid immunoreactivity was quantified by RIA (1/1000 of total sample) by using the SHO-3 antibody after incubations with sad (filled circles) or GFP (open circles). UV absorption was measured at 248 nm (solid line). (Inset, TLC) TLC/RIA (chloroform/ethanol) of RP-HPLC-purified ecdysone (E) product (1/1000 of total sample). (Inset, MS) RP-HPLC/ESI-MS on an LCQDECA (Thermo Finnigan, San Jose, CA) of the TLC-purified E product.
Fig 4.
RP-HPLC/TLC analysis of ecdysteroids after _sad_-, _dib_-, sad and _dib_-, or GFP-transfected S2 cell incubations with the [3H]ketotriol (2,22dE) substrate. (A) sad (filled circles) or GFP (open circles) + [3H]ketotriol. ?, Unidentified metabolite conjugate. (Inset) TLC analysis of [3H[22dE product. (B) dib (filled circles) or GFP (open circles) + [3H]ketotriol. (Inset) TLC of [3H]2dE product. (C) sad and dib (filled circles) or GFP (open circles) + [3H]ketotriol. (Inset) TLC of [3H]E (ecdysone) product.
References
- Gilbert L. I., Rybczynski, R. & Warren, J. T. (2002) Annu. Rev. Entomol. 47**,** 883-916. - PubMed
- Henrich V. C., Rybczynski, R. & Gilbert, L. I. (1999) in Vitamins and Hormones, ed. Litwack, G. (Academic, San Diego), Vol. 55, pp. 73–125. - PubMed
- Thummel C. S. (1995) Cell 83**,** 871-877. - PubMed
- Kappler C., Kabbouh, M., Hetru, C., Durst, F. & Hoffmann, J. (1988) J. Steroid Biochem. 31**,** 891-898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES10126/ES/NIEHS NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- HD33692/HD/NICHD NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- P30 CA16086/CA/NCI NIH HHS/United States
- K12 HD033692/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous