Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags - PubMed (original) (raw)
. 2002 Aug 20;99(17):11049-54.
doi: 10.1073/pnas.172170199. Epub 2002 Aug 12.
Ljiljana Pasa-Tolic', Gordon A Anderson, David J Anderson, Deanna L Auberry, John R Battista, Michael J Daly, Jim Fredrickson, Kim K Hixson, Heather Kostandarithes, Christophe Masselon, Lye Meng Markillie, Ronald J Moore, Margaret F Romine, Yufeng Shen, Eric Stritmatter, Nikola Tolic', Harold R Udseth, Amudhan Venkateswaran, Kwong-Kwok Wong, Rui Zhao, Richard D Smith
Affiliations
- PMID: 12177431
- PMCID: PMC129300
- DOI: 10.1073/pnas.172170199
Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags
Mary S Lipton et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Understanding biological systems and the roles of their constituents is facilitated by the ability to make quantitative, sensitive, and comprehensive measurements of how their proteome changes, e.g., in response to environmental perturbations. To this end, we have developed a high-throughput methodology to characterize an organism's dynamic proteome based on the combination of global enzymatic digestion, high-resolution liquid chromatographic separations, and analysis by Fourier transform ion cyclotron resonance mass spectrometry. The peptides produced serve as accurate mass tags for the proteins and have been used to identify with high confidence >61% of the predicted proteome for the ionizing radiation-resistant bacterium Deinococcus radiodurans. This fraction represents the broadest proteome coverage for any organism to date and includes 715 proteins previously annotated as either hypothetical or conserved hypothetical.
Figures
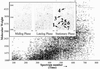
Fig 1.
Two-dimensional display of a capillary LC-FTICR analysis in which >50,000 putative polypeptides were detected from a tryptic digest of D. radiodurans proteins harvested in mid-log phase (OD600 0.3–0.4; 30°C). Inset shows portions of displays for peptides from D. radiodurans harvested in mid-log (Left), late-log (Center), and poststationary (Right) phases (spot size reflects relative abundance). Each individual spot corresponds to a peptide that can be identified along with the parent protein using AMTs. Spot identifications: 1: DR1314 (hypothetical protein); 2: DR1790 (homolog of yellow/royal jelly protein of insects); 3: DR1314 (hypothetical protein); 4: DR0309 (elongation factor Tu); 5: DR2577 (S-layer protein); 6: DR0989 (membrane protein); and 7: DR1124 (S-layer protein).
Fig 2.
(A) The qualitative pattern of ORF expression detected using AMTs for various conditions by TIGR assigned functional category (2) (red, AMTs detected; green, AMTs not observed). Each culture condition (as described in Experimental Protocols) was analyzed at least two times. (B) Expansion of the pattern for predicted hypothetical proteins, illustrating similarities and differences under different culture conditions. (C) Expansion showing several representative hypothetical proteins.
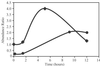
Fig 3.
Change in relative abundance of (•) RecA and (♦) DNA-directed RNA polymerase in cells after exposure to 17.5 kGy of ionizing radiation obtained be analysis of a mixture of cells grown on unlabeled media and the 15N-labeled reference proteome (control, nonirradiated cells). Cells were prepared at 0, 3, 7, 9, and 12 hr after exposure.
Comment in
- New technology may reveal mechanisms of radiation resistance in Deinococcus radiodurans.
Mrazek J. Mrazek J. Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):10943-4. doi: 10.1073/pnas.182429699. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177454 Free PMC article. No abstract available.
Similar articles
- The use of accurate mass tags for high-throughput microbial proteomics.
Smith RD, Anderson GA, Lipton MS, Masselon C, Pasa-Tolic L, Shen Y, Udseth HR. Smith RD, et al. OMICS. 2002;6(1):61-90. doi: 10.1089/15362310252780843. OMICS. 2002. PMID: 11881835 Review. - An accurate mass tag strategy for quantitative and high-throughput proteome measurements.
Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. Smith RD, et al. Proteomics. 2002 May;2(5):513-23. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. Proteomics. 2002. PMID: 11987125 - iTRAQ-based proteomic analysis of Deinococcus radiodurans in response to 12C6+ heavy ion irradiation.
Gao Y, Li N, Zhou Y, Zhang Z, Zhang Y, Fan P, Zhou H, Zhang T, Chang L, Gao H, Li Y, Kang X, Xie Q, Lyu Z, Xu P. Gao Y, et al. BMC Microbiol. 2022 Nov 4;22(1):264. doi: 10.1186/s12866-022-02676-x. BMC Microbiol. 2022. PMID: 36333788 Free PMC article. - Advanced mass spectrometric methods for the rapid and quantitative characterization of proteomes.
Smith RD. Smith RD. Comp Funct Genomics. 2002;3(2):143-50. doi: 10.1002/cfg.159. Comp Funct Genomics. 2002. PMID: 18628837 Free PMC article. - DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans.
Minton KW. Minton KW. Mol Microbiol. 1994 Jul;13(1):9-15. doi: 10.1111/j.1365-2958.1994.tb00397.x. Mol Microbiol. 1994. PMID: 7984097 Review.
Cited by
- Automated, reproducible, titania-based phosphopeptide enrichment strategy for label-free quantitative phosphoproteomics.
Richardson BM, Soderblom EJ, Thompson JW, Moseley MA. Richardson BM, et al. J Biomol Tech. 2013 Apr;24(1):8-16. doi: 10.7171/jbt.13-2401-002. J Biomol Tech. 2013. PMID: 23542237 Free PMC article. - Thylakoid membrane proteomics.
Whitelegge JP. Whitelegge JP. Photosynth Res. 2003;78(3):265-77. doi: 10.1023/B:PRES.0000006828.65688.0d. Photosynth Res. 2003. PMID: 16245055 - DRJAMM Is Involved in the Oxidative Resistance in Deinococcus radiodurans.
Cai J, Pan C, Zhao Y, Xu H, Tian B, Wang L, Hua Y. Cai J, et al. Front Microbiol. 2022 Jan 28;12:756867. doi: 10.3389/fmicb.2021.756867. eCollection 2021. Front Microbiol. 2022. PMID: 35154022 Free PMC article. - The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle.
Welsh EA, Liberton M, Stöckel J, Loh T, Elvitigala T, Wang C, Wollam A, Fulton RS, Clifton SW, Jacobs JM, Aurora R, Ghosh BK, Sherman LA, Smith RD, Wilson RK, Pakrasi HB. Welsh EA, et al. Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):15094-9. doi: 10.1073/pnas.0805418105. Epub 2008 Sep 23. Proc Natl Acad Sci U S A. 2008. PMID: 18812508 Free PMC article. - PprM is necessary for up-regulation of katE1, encoding the major catalase of Deinococcus radiodurans, under unstressed culture conditions.
Jeong SW, Seo HS, Kim MK, Choi JI, Lim HM, Lim S. Jeong SW, et al. J Microbiol. 2016 Jun;54(6):426-31. doi: 10.1007/s12275-016-6175-8. Epub 2016 May 27. J Microbiol. 2016. PMID: 27225459
References
- Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1325. - PubMed
- Daly M. J. (2000) Biotechnology 11, 280-285. - PubMed
- Smith K. C. & Martignoni, K. D. (1976) Photochem. Photobiol. 24, 515-523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources