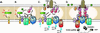Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies - PubMed (original) (raw)
Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies
Doyle V Ward et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Numerous bacterial pathogens use type IV secretion systems (T4SS) to deliver virulence factors directly to the cytoplasm of plant, animal, and human host cells. Here, evidence for interactions among components of the Agrobacterium tumefaciens vir-encoded T4SS is presented. The results derive from a high-resolution yeast two-hybrid assay, in which a library of small peptide domains of T4SS components was screened for interactions. The use of small peptides overcomes problems associated with assaying for interactions involving membrane-associated proteins. We established interactions between VirB11 (an inner membrane pore-forming protein), VirB9 (a periplasmic protein), and VirB7 (an outer membrane-associated lipoprotein and putative pilus component). We provide evidence for an interaction pathway, among conserved members of a T4SS, spanning the A. tumefaciens envelope and including a potential pore protein. In addition, we have determined interactions between VirB1 (a lytic transglycosylase likely involved in the local remodeling of the peptidoglycan) and primarily VirB8, but also VirB4, VirB10, and VirB11 (proteins likely to assemble the core structure of the T4SS). VirB4 interacts with VirB8, VirB10, and VirB11, also establishing a connection to the core components. The identification of these interactions suggests a model for assembly of the T4SS.
Figures
Fig 1.
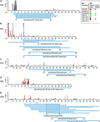
Minimal interacting domains of VirB7 and VirB9 proteins. The N-terminal residues of VirB9 and VirB7 peptide prey isolated from the high-resolution library were determined. The position of the peptide termini are plotted versus the codon position of the full-length prey protein (x axis) and the number of isolates identified (y axis). The lines below each plot represent the minimal prey domain determined to interact with the bait used. (A) VirB9 prey isolated with full-length VirB7 bait. (B) Total VirB7 prey isolated with two VirB9 baits, VirB9(14–293) and VirB9(183–293).
Fig 2.
Minimal interacting domains of VirB proteins. The N-terminal and C-terminal residues of prey isolated from the high-resolution library are plotted versus codon position (x axis) and the number of isolates identified (y axis). Data are color-coded for each bait used (see Inset). Prey that have determined N and C termini and that were isolated by a unique bait are aligned to define regions in common, indicating minimal interacting domains (horizontal blue lines below the amino acid numbers). The prey represented are as follows VirB9 (A), VirB1 (B), VirB4 (C), VirB8 (D), and VirB10 (E).
Fig 3.
VirB protein interactions. The pairwise interactions identified among the VirB proteins are summarized. The horizontal lines represent the peptide sequence of each VirB protein with the length in amino acids indicated to the right. Blue shaded regions indicate the minimal domains that contribute to each pairwise interaction. The numbers indicate the specific residue in the coding sequence that delimits each domain. The heavy arrows indicate the direction in which the interaction was observed, from bait to prey. The data most easily sort into two main complexes; the first consists of VirB1, VirB7, VirB9, and VirB11, and the second consists of VirB1, VirB4, VirB8, VirB10, and VirB11. VirB4–VirB4 interactions are presented as a third group. VirB1–VirB1, VirB1–VirB4, VirB8–VirB8, VirB8–VirB10, and VirB10–VirB11 interactions are not represented but are addressed in the text.
Fig 4.
Model for assembly of the Agrobacterium T4SS. Protein interactions determined in this work are depicted in A and B. Previously determined interactions are shown in C. The double lines at the top and bottom of each panel represent the inner and outer membranes, and the shaded region represents the periplasmic space and peptidoglycan. (A) VirB8 functions as a “founding member” of the T4SS and recruits VirB1 to the site of assembly. VirB1 locally remodels the peptidoglycan (represented by decreased shading of the periplasm). (B) VirB1 activity allows recruitment of other T4SS components such as VirB4, VirB7, VirB9, VirB10, and VirB11 by clearing the peptidoglycan or by recruiting components via direct interactions. VirB7–VirB9 heterodimers are critical for stability of T4SS proteins. (C) As the assembly matures, the remaining components are recruited, including VirB3, the pilus (VirB2, VirB5), and inner membrane components (VirB6, VirD4). VirB1* may function extracellularly, and its loose association with the surface is indicated. VirB7 and VirB5 contribute to pilus assembly. VirB4, VirB6, VirB11, and VirD4 all have been postulated to create channels for substrate secretion (blue arrows). (D) A model for assembled T4SS based on interactions described in A–C. VirB7–11 and VirB4 form a functional core of the T4SS that spans both bacterial membranes. Known substrates for export include VirE2 and, perhaps, its specific chaperone VirE1, VirF, and the T-complex (VirD2–ssDNA). VirD4 is likely the recognition protein coupling VirD2 export to the T4SS and may function as a DNA-helicase to liberate the T-strand from the Ti plasmid. VirB11 forms a pore and may facilitate substrate export.
Similar articles
- Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens.
Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. Krall L, et al. Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11405-10. doi: 10.1073/pnas.172390699. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177443 Free PMC article. - VirB8: a conserved type IV secretion system assembly factor and drug target.
Baron C. Baron C. Biochem Cell Biol. 2006 Dec;84(6):890-9. doi: 10.1139/o06-148. Biochem Cell Biol. 2006. PMID: 17215876 Review. - Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8.
Kumar RB, Das A. Kumar RB, et al. J Bacteriol. 2001 Jun;183(12):3636-41. doi: 10.1128/JB.183.12.3636-3641.2001. J Bacteriol. 2001. PMID: 11371528 Free PMC article. - Exploring cargo transport mechanics in the type IV secretion systems.
Li J, Wolf SG, Elbaum M, Tzfira T. Li J, et al. Trends Microbiol. 2005 Jul;13(7):295-8. doi: 10.1016/j.tim.2005.05.002. Trends Microbiol. 2005. PMID: 15923116 Review.
Cited by
- Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates.
Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. Morrison SS, et al. PLoS One. 2012;7(5):e37553. doi: 10.1371/journal.pone.0037553. Epub 2012 May 25. PLoS One. 2012. PMID: 22662170 Free PMC article. - The versatile bacterial type IV secretion systems.
Cascales E, Christie PJ. Cascales E, et al. Nat Rev Microbiol. 2003 Nov;1(2):137-49. doi: 10.1038/nrmicro753. Nat Rev Microbiol. 2003. PMID: 15035043 Free PMC article. Review. - Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli.
Zahrl D, Wagner M, Bischof K, Koraimann G. Zahrl D, et al. J Bacteriol. 2006 Sep;188(18):6611-21. doi: 10.1128/JB.00632-06. J Bacteriol. 2006. PMID: 16952953 Free PMC article. - Proteomic profiling of the outer membrane fraction of the obligate intracellular bacterial pathogen Ehrlichia ruminantium.
Moumène A, Marcelino I, Ventosa M, Gros O, Lefrançois T, Vachiéry N, Meyer DF, Coelho AV. Moumène A, et al. PLoS One. 2015 Feb 24;10(2):e0116758. doi: 10.1371/journal.pone.0116758. eCollection 2015. PLoS One. 2015. PMID: 25710494 Free PMC article. - Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori.
Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Terradot L, et al. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4596-601. doi: 10.1073/pnas.0408927102. Epub 2005 Mar 11. Proc Natl Acad Sci U S A. 2005. PMID: 15764702 Free PMC article.
References
- Linton K. J. & Higgins, C. F. (1998) Mol. Microbiol. 28, 5-13. - PubMed
- Sandkvist M. (2001) Mol. Microbiol. 40, 271-283. - PubMed
- Plano G. V., Day, J. B. & Ferracci, F. (2001) Mol. Microbiol. 40, 284-293. - PubMed
- Jacob-Dubuisson F., Locht, C. & Antoine, R. (2001) Mol. Microbiol. 40, 306-313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources