Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3 - PubMed (original) (raw)
Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3
Jianhe Huang et al. J Biol Chem. 2002.
Abstract
Hypoxia-inducible factor (HIF) is a heterodimeric transcription factor induced by hypoxia. Under normoxic conditions, site-specific proline hydroxylation of the alpha subunits of HIF allows recognition by the von Hippel-Lindau tumor suppressor protein (VHL), a component of an E3 ubiquitin ligase complex that targets these subunits for degradation by the ubiquitin-proteasome pathway. Under hypoxic conditions, this hydroxylation is inhibited, allowing the alpha subunits of HIF to escape VHL-mediated degradation. Three enzymes, prolyl hydroxylase domain-containing proteins 1, 2, and 3 (PHD1, -2, and -3; also known as HIF prolyl hydroxylase 3, 2, and 1, respectively), have recently been identified that catalyze proline hydroxylation of HIF alpha subunits. These enzymes hydroxylate specific prolines in HIF alpha subunits in the context of a strongly conserved LXXLAP sequence motif (where X indicates any amino acid and P indicates the hydroxylacceptor proline). We report here that PHD2 has the highest specific activity toward the primary hydroxylation site of HIF-1alpha. Furthermore, and unexpectedly, mutations can be tolerated at the -5, -2, and -1 positions (relative to proline) of the LXXLAP motif. Thus, these results provide evidence that the only obligatory residue for proline hydroxylation in HIF-1alpha is the hydroxylacceptor proline itself.
Figures
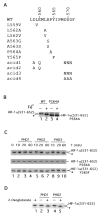
Fig. 1. PHD-induced mobility shift of HIF-1α-(531–652)
A, sequence of HIF-1α from residue 557–572. Wild type sequence is shown at the top, and mutants are indicated below. B, reticulocyte lysate-induced mobility shift of HIF-1α-(531–652). 35S-labeled GAL4-HIF-1α-(531–652) or GAL4-HIF-1α-(531–652)-P564A was prepared by in vitro transcription and translation with reticulocyte lysates in the absence or presence of 100 μM FeCl2, as shown. Products were subjected to SDS-PAGE and autoradiography. C, PHD-induced mobility shift of HIF-1α-(531–652). In vitro translated, 35S-labeled wild type (top panel), P564A (middle panel), or Y565F (bottom panel) GAL4-HIF-1α-(531–652) was prepared using wheat germ extracts and incubated with 0.5 μg of His6-FLAG-PHD1, His6-FLAG-PHD2, or His6-FLAG-PHD3 for the indicated times at 30 °C. The reaction products were subjected to SDS-PAGE and autoradiography. One of four representative results is shown. D, 2-oxoglutarate dependence of PHD-induced mobility shift. His6-FLAG-PHD1 or His6-FLAG-PHD2 was incubated with 35S-labeled, in vitro translated GAL4-HIF-1α-(531–652) for 1 h at 30 °C in the absence or presence of 1 mM 2-oxoglutarate. Reaction products were subjected to SDS-PAGE and autoradiography. B–D, the positions of the in vitro translated proteins are indicated, and asterisks indicate more rapidly migrating species (see text).
Fig. 2. PHD-induced hydroxylation of wild type and mutant HIF-1α-(531–652)
A, in vitro translated, 35S-labeled wild type or mutant GAL4-HIF-1α-(531–652) was incubated with 0.5 μg of His6-PHD1, His6-PHD2, or His6-PHD3 for the indicated times. Hydroxylated reaction products were then isolated by first incubating with His6-FLAG-VBC and then by immunoprecipitating with anti-FLAG antibodies coupled to agarose. The immunoprecipitated reaction products were subjected to SDS-PAGE and autoradiography. B, same as in A except that His6-PHD2 and either HA-HIF-1α or GAL4-HIF-1α-(531–652) were employed. Input (In) designates 20% of the substrate analyzed at a given time point. One of three representative results is shown.
Fig. 3. Interaction between PHDs and HIF-1α
A, PHD pull-down analysis. In vitro translated, 35S-labeled wild type or mutant GAL4-HIF-1α-(531–652) prepared with wheat germ extracts were incubated with 1 μg of His6-FLAG-PHD1, His6-FLAG-PHD2, or His6-FLAG-PHD3. The PHDs were then immunoprecipitated with anti-FLAG antibodies coupled to agarose. Coimmunoprecipitating wild type or mutant GAL4-HIF-1α-(531–652) was then subjected to SDS-PAGE and autoradiography. B, in vitro translated, 35S-labeled FLAG-PHD1, FLAG-PHD2, or FLAG-PHD3 was incubated with GST or GST-HIF-1α-(531–575) prebound to GSH-agarose. After washing, bound PHD was subjected to SDS-PAGE and autoradiography. The positions of the PHDs are shown to the left, and those of molecular weight markers are to the right. In A and B, Input designates 10% of input.
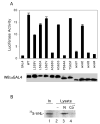
Fig. 4
A, dominant negative behavior of wild type and mutant HIF-1α-(531–652). COS-1 cells were cotransfected with 0.5 μg of (eHRE)3-Luc, 0.05 μg of pRL-TK (Promega), and 0.5 μg of wild type or mutant pcDNA3-GAL4-HIF-1α-(531–652), or pcDNA3-GAL4. Cellular extracts were assayed for luciferase activity, normalizing to that of the Renilla luciferase internal transfection control. One of three representative results, performed in triplicate, is shown. Aliquots of the extracts were analyzed by Western blotting with anti-GAL4 antibodies (bottom). B, stimulus-sensitive endogenous PHD activity. COS-1 cells were mock (N) or cobalt (Co++)-treated for 2 h. Cellular lysates were prepared, and endogenous PHD from these lysates was then isolated by incubation with GST-HIF-1α-(531–575) immobilized on GSH-agarose. Hydroxylation reactions were performed on the resins, and the extent of HIF-1α-(531–575) hydroxylation was then assayed by subsequent incubation with 35S-labeled, in vitro translated VHL. Bound VHL was subjected to SDS-PAGE and autoradiography. In a control reaction, lysates were omitted. The position of VHL is shown, whereas “In ” designates 10% of the input VHL. One of two representative results is shown.
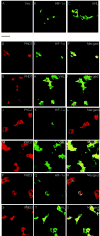
Fig. 5. Cellular localization of PHDs, HIF-1α, and VHL in transfected COS-1 cells
COS-1 cells were cotransfected with constructs encoding HA-HIF-1α (B, D–F, J–L, and P–R) or HA-VHL (C, G–I, M–O, and S–U) with or without ones for FLAG-tagged PHD1 (D–I), PHD2 (J–O), or PHD3 (P–U), or pcDNA3 alone (A). At 20-h post-transfection, the cells were fixed, permeabilized, and stained with anti-FLAG or anti-HA antibodies, or both. Confocal images of single color and an overlay of two colors (Merged) are shown. The bar between panels A and D indicates 50 μM.
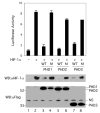
Fig. 6. Inhibition of HIF-1α transcriptional activity by PHD
COS-1 cells were cotransfected with 0.25 μg of (eHRE)3-Luc, 0.025 μg of pRL-TK (Promega), 0.5 μg of pcDNA3-HA-HIF-1α, and 0.1 μg of pcDNA3-FLAG-PHD1, pcDNA3-FLAG-PHD1 (H297A), pcDNA3-FLAG-PHD2, pcDNA3-FLAG-PHD2 (R383A), pcDNA3-FLAG-PHD3, or pcDNA3-FLAG-PHD3 (H135A). The total DNA dose was adjusted to 0.875 μg with pcDNA3. Cellular extracts were assayed for luciferase activity, normalizing to that of the Renilla luciferase internal transfection control. One of three representative results, performed in triplicate, is shown. Aliquots of extracts were also subjected to Western blotting with anti-HIF-1α or anti-FLAG antibodies. The positions of the PHDs and a nonspecific (NS) band are shown to the right, and those of molecular weight markers are to the left. WT and M indicate the wild type and mutant forms of PHD described above, respectively.
Similar articles
- Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases.
Marxsen JH, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, Jelkmann W, Jaakkola P, Metzen E. Marxsen JH, et al. Biochem J. 2004 Aug 1;381(Pt 3):761-7. doi: 10.1042/BJ20040620. Biochem J. 2004. PMID: 15104534 Free PMC article. - HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia.
Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J. Berra E, et al. EMBO J. 2003 Aug 15;22(16):4082-90. doi: 10.1093/emboj/cdg392. EMBO J. 2003. PMID: 12912907 Free PMC article. - Leu-574 of human HIF-1alpha is a molecular determinant of prolyl hydroxylation.
Kageyama Y, Koshiji M, To KK, Tian YM, Ratcliffe PJ, Huang LE. Kageyama Y, et al. FASEB J. 2004 Jun;18(9):1028-30. doi: 10.1096/fj.03-1233fje. Epub 2004 Apr 14. FASEB J. 2004. PMID: 15084514 - Hypoxia-inducible factor 1 (HIF-1) pathway.
Semenza GL. Semenza GL. Sci STKE. 2007 Oct 9;2007(407):cm8. doi: 10.1126/stke.4072007cm8. Sci STKE. 2007. PMID: 17925579 Review. - Proline hydroxylation and gene expression.
Kaelin WG. Kaelin WG. Annu Rev Biochem. 2005;74:115-28. doi: 10.1146/annurev.biochem.74.082803.133142. Annu Rev Biochem. 2005. PMID: 15952883 Review.
Cited by
- Identification of Small-Molecule PHD2 Zinc Finger Inhibitors that Activate Hypoxia Inducible Factor.
Arsenault PR, Song D, Bergkamp M, Ravaschiere AM, Navalsky BE, Lieberman PM, Lee FS. Arsenault PR, et al. Chembiochem. 2016 Dec 14;17(24):2316-2323. doi: 10.1002/cbic.201600493. Epub 2016 Nov 11. Chembiochem. 2016. PMID: 27770548 Free PMC article. - A common polymorphism in the oxygen-dependent degradation (ODD) domain of hypoxia inducible factor-1alpha (HIF-1alpha) does not impair Pro-564 hydroxylation.
Percy MJ, Mooney SM, McMullin MF, Flores A, Lappin TR, Lee FS. Percy MJ, et al. Mol Cancer. 2003 Sep 9;2:31. doi: 10.1186/1476-4598-2-31. Mol Cancer. 2003. PMID: 14521712 Free PMC article. - Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous?
Strowitzki MJ, Cummins EP, Taylor CT. Strowitzki MJ, et al. Cells. 2019 Apr 26;8(5):384. doi: 10.3390/cells8050384. Cells. 2019. PMID: 31035491 Free PMC article. Review. - Identification of prolyl hydroxylation modifications in mammalian cell proteins.
Arsenault PR, Heaton-Johnson KJ, Li LS, Song D, Ferreira VS, Patel N, Master SR, Lee FS. Arsenault PR, et al. Proteomics. 2015 Apr;15(7):1259-67. doi: 10.1002/pmic.201400398. Epub 2015 Jan 19. Proteomics. 2015. PMID: 25421965 Free PMC article. - Endothelial EGLN3-PKM2 signaling induces the formation of acute astrocytic barrier to alleviate immune cell infiltration after subarachnoid hemorrhage.
Duan M, Ru X, Zhou J, Li Y, Guo P, Kang W, Li W, Chen Z, Feng H, Chen Y. Duan M, et al. Fluids Barriers CNS. 2024 May 16;21(1):42. doi: 10.1186/s12987-024-00550-8. Fluids Barriers CNS. 2024. PMID: 38755642 Free PMC article.
References
- Semenza GL. Curr Opin Cell Biol. 2001;13:167–171. - PubMed
- Maxwell PH, Pugh CW, Ratcliffe PJ. Adv Exp Med Biol. 2001;502:365–376. - PubMed
- Kaelin WG., Jr Genes Dev. 2002;16:1441–1445. - PubMed
- Semenza GL. Annu Rev Cell Dev Biol. 1999;15:551–578. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases